[Paper review] Unraveling the Genetic Complexity of Coronary Artery Disease: A New Approach Linking Variants to Genes to Pathways
In a groundbreaking study published in Nature, Schnitzler et al. introduce a novel approach called Variant-to-Gene-to-Program (V2G2P) to systematically link genetic variants associated with complex diseases to causal genes and cellular pathways. By integrating genome-wide association study (GWAS) data, epigenomic annotations, and single-cell perturbation screens (Perturb-seq), the researchers uncover new insights into the genetic basis of coronary artery disease (CAD) and demonstrate the power of their approach to identify novel disease mechanisms and potential therapeutic targets.
The V2G2P approach, applied to CAD, reveals the convergence of multiple risk loci on endothelial cell transcriptional programs related to the cerebral cavernous malformation (CCM) signaling pathway. The study identifies two new CAD genes, CCM2 and TLNRD1, which interact physically and functionally to regulate atherosclerosis risk through the CCM pathway. These findings suggest that decreased CCM signaling in endothelial cells may be protective against CAD, contrasting with its role in the rare monogenic disease cerebral cavernous malformation.
This work represents a significant advance in our understanding of the genetic complexity of common diseases and provides a framework for future investigations into the molecular basis of human health and disease. The V2G2P approach has the potential to accelerate the discovery of novel therapeutic targets and guide the development of precision medicine strategies for a wide range of complex disorders.
Introduction
Genome-wide association studies (GWAS) have identified hundreds of genetic variants associated with complex diseases, such as coronary artery disease (CAD). However, linking these variants to the genes and biological pathways they influence remains a major challenge. Understanding the mechanistic links between genetic risk factors and disease is crucial for developing targeted therapies and interventions.
To address this challenge, Schnitzler et al. introduced a novel approach called Variant-to-Gene-to-Program (V2G2P) in their recent paper published in Nature. This method systematically links disease-associated genetic variants to causal genes and cellular pathways by integrating epigenomic data, single-cell RNA sequencing, and CRISPR-based perturbations.
The V2G2P approach offers an unbiased and comprehensive way to identify the convergence of risk variants on specific disease mechanisms. By applying this method to CAD, the researchers uncovered new insights into the role of endothelial cells and the CCM signaling pathway in the development of atherosclerosis.
In this blog post, we will delve into the details of the V2G2P approach, its application to CAD, and the significant implications of this work for understanding complex disease genetics and guiding future therapeutic strategies.
The V2G2P Approach
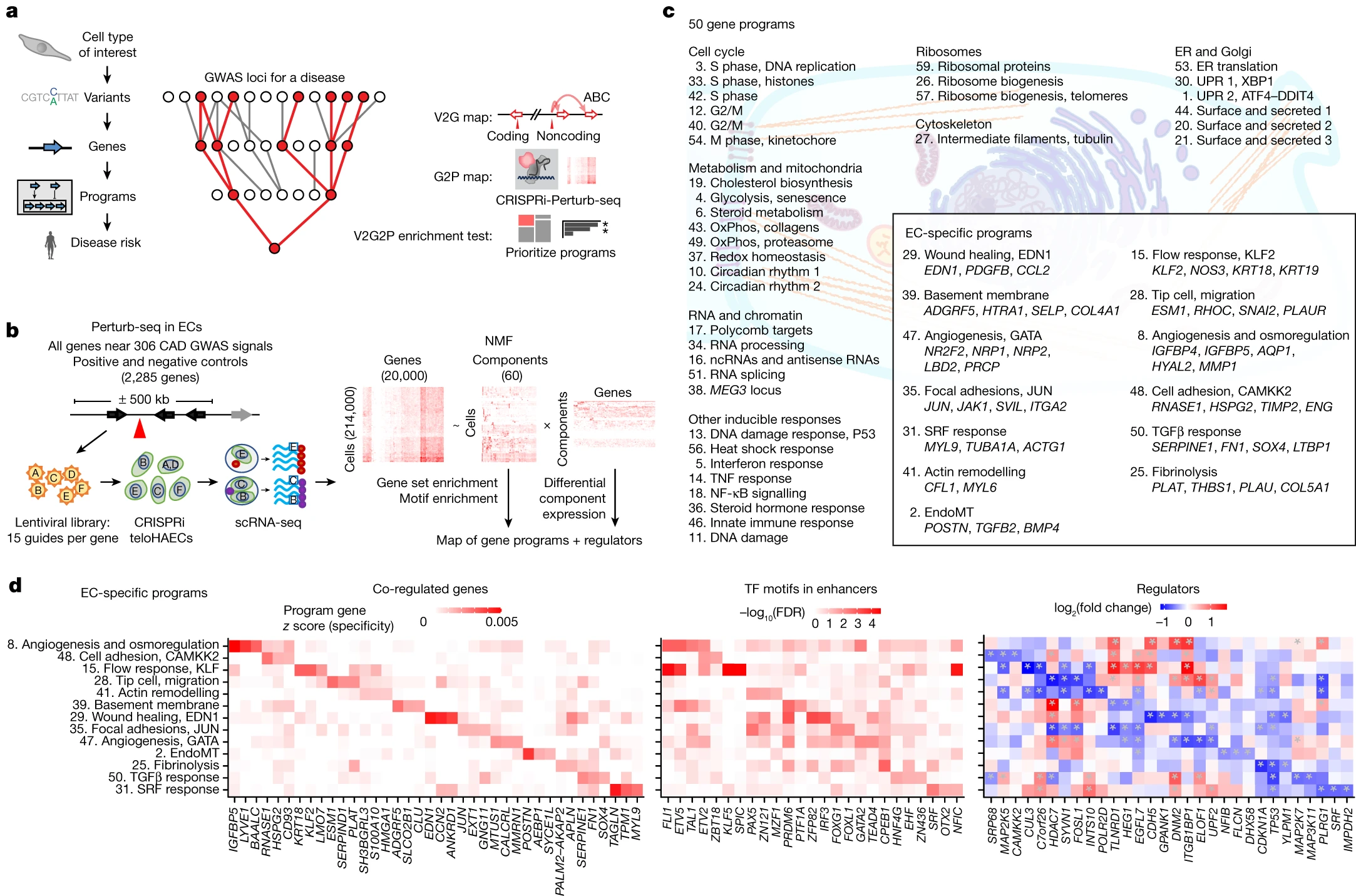
The V2G2P approach is a five-step process that integrates various genomic and functional data to systematically link disease-associated variants to causal genes and cellular pathways. Let’s break down each step in detail.
Step 1: Identifying the Relevant Cell Type
The first step in the V2G2P approach is to identify the cell type most relevant to the disease of interest. This is done by examining the enrichment of disease risk variants in cell-type-specific enhancers. In the case of CAD, Schnitzler et al. focused on endothelial cells, as they found a significant enrichment of CAD risk variants in endothelial cell enhancers.
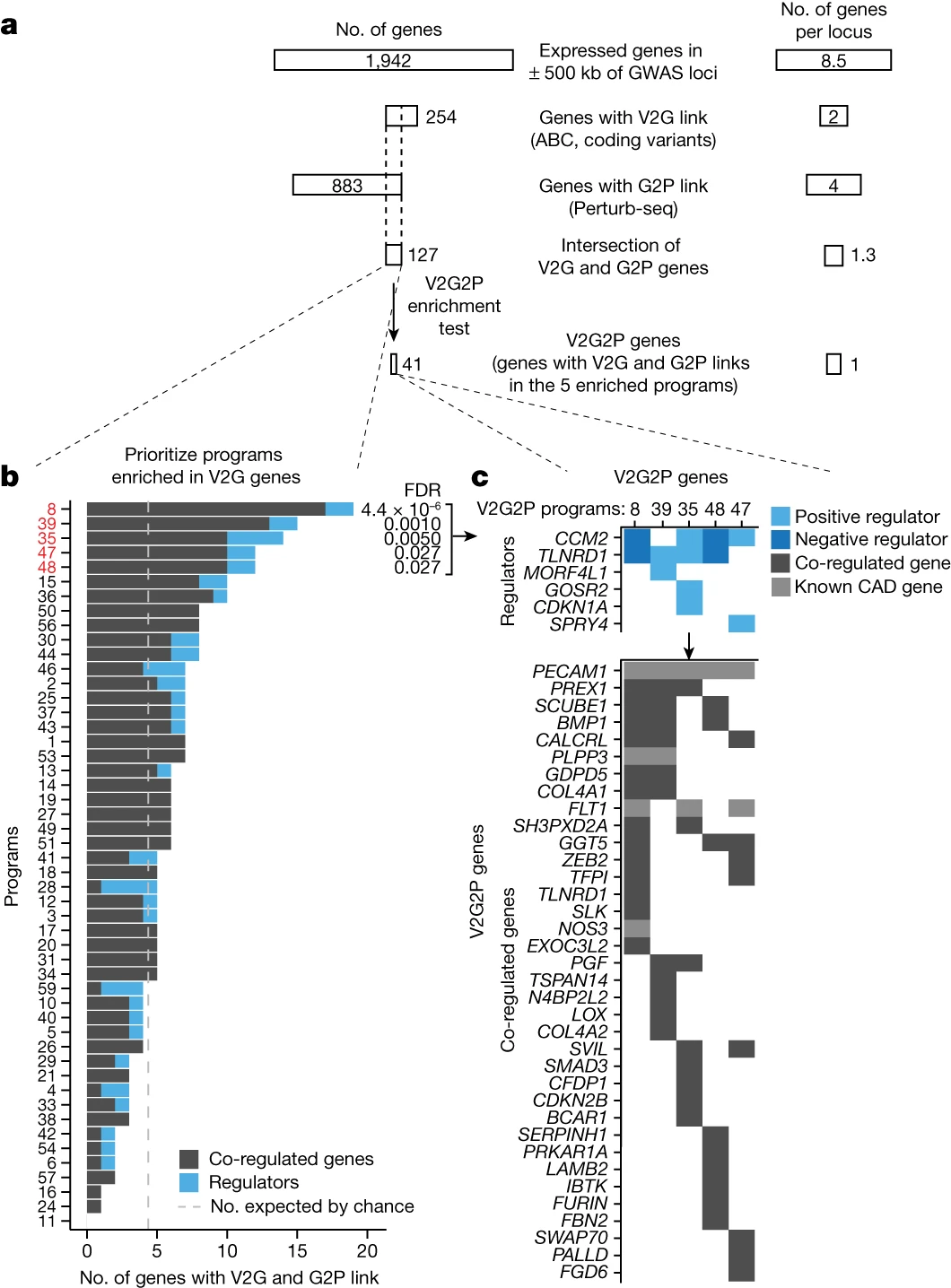
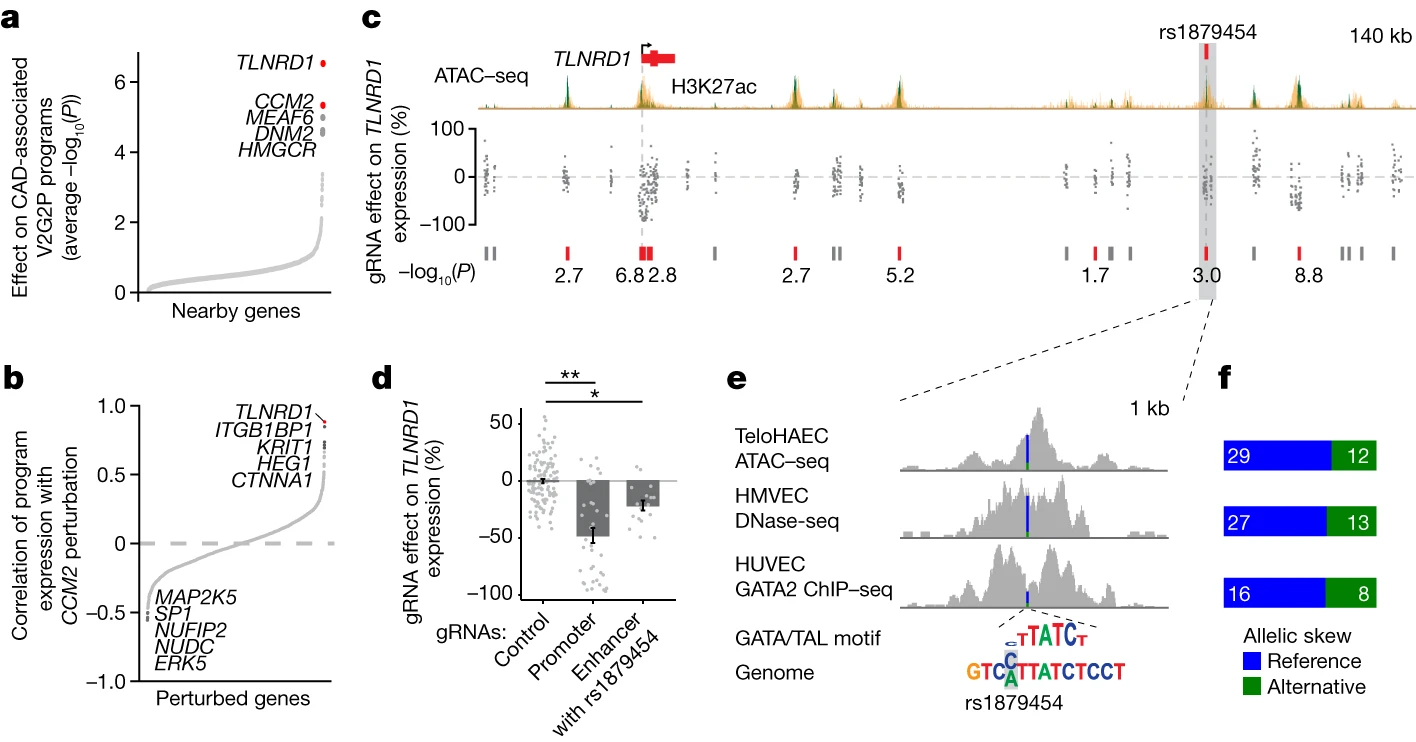
Step 2: Building a Variant-to-Gene (V2G) Map (Fig. 1b)
The next step involves constructing a map that links disease-associated variants to potential target genes using epigenomic data in the relevant cell type. The researchers used information from endothelial cell enhancers, coding regions, and splice sites to establish these variant-to-gene links.
Step 3: Building a Gene-to-Program (G2P) Map with Perturb-seq (Fig. 1c)
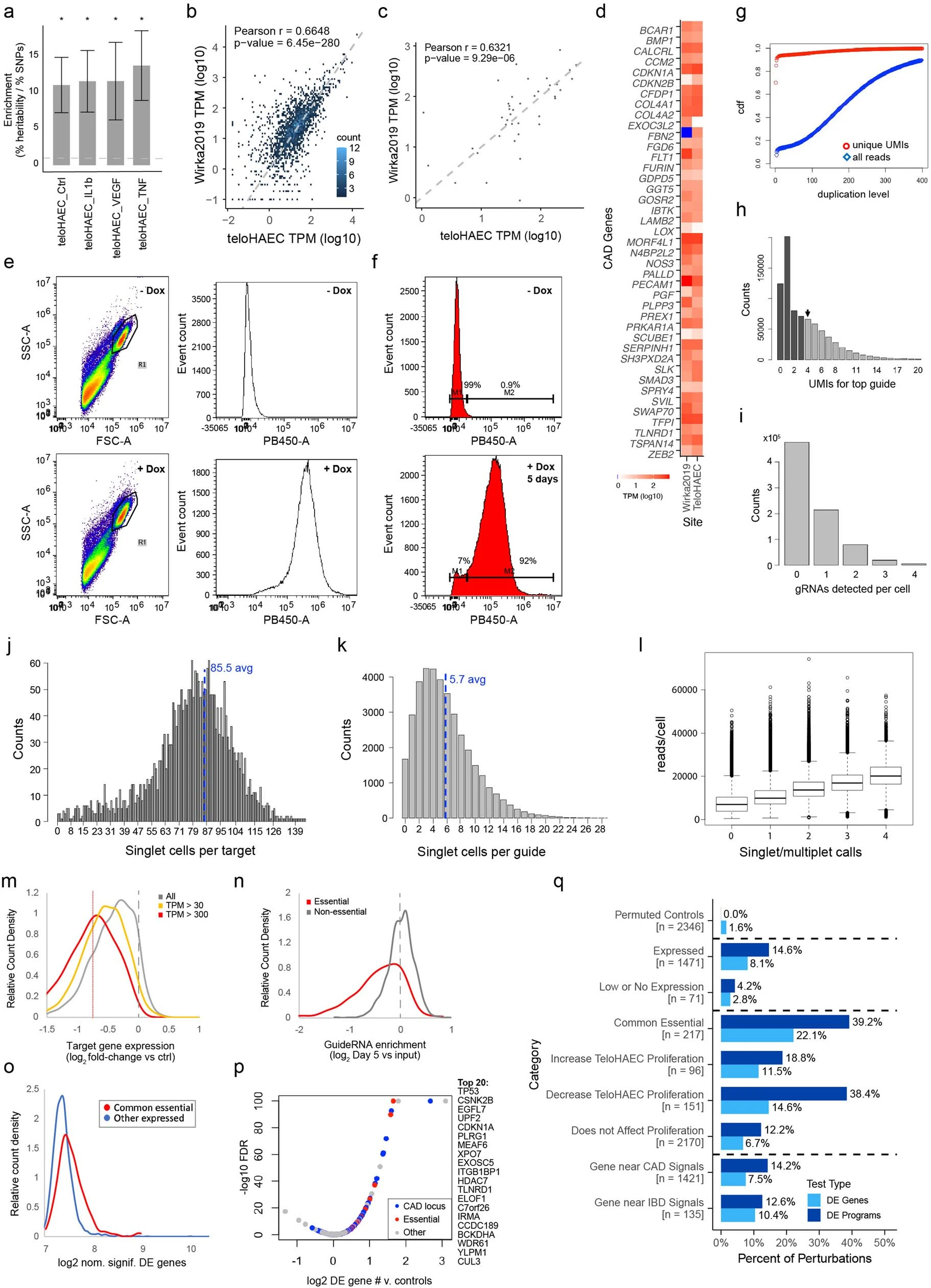
To identify sets of co-regulated genes (“programs”) and link them to upstream regulator genes, the researchers employed a technique called Perturb-seq. They systematically knocked down all expressed genes within ±500 kb of the CAD GWAS signals using CRISPR interference (CRISPRi) and measured the effects on gene expression using single-cell RNA sequencing (scRNA-seq). They then applied unsupervised machine learning to define gene programs without relying on prior knowledge of gene sets or pathways.
Step 4: Identifying Disease-Associated Programs
To determine which gene programs are associated with the disease of interest, the researchers developed a statistical test called V2G2P enrichment. This test assesses whether the genes linked to risk variants (from the V2G map) are enriched in specific programs (from the G2P map). In the CAD study, they identified five endothelial cell programs that were significantly enriched for genes linked to CAD risk variants.
Step 5: Studying Prioritized Genes
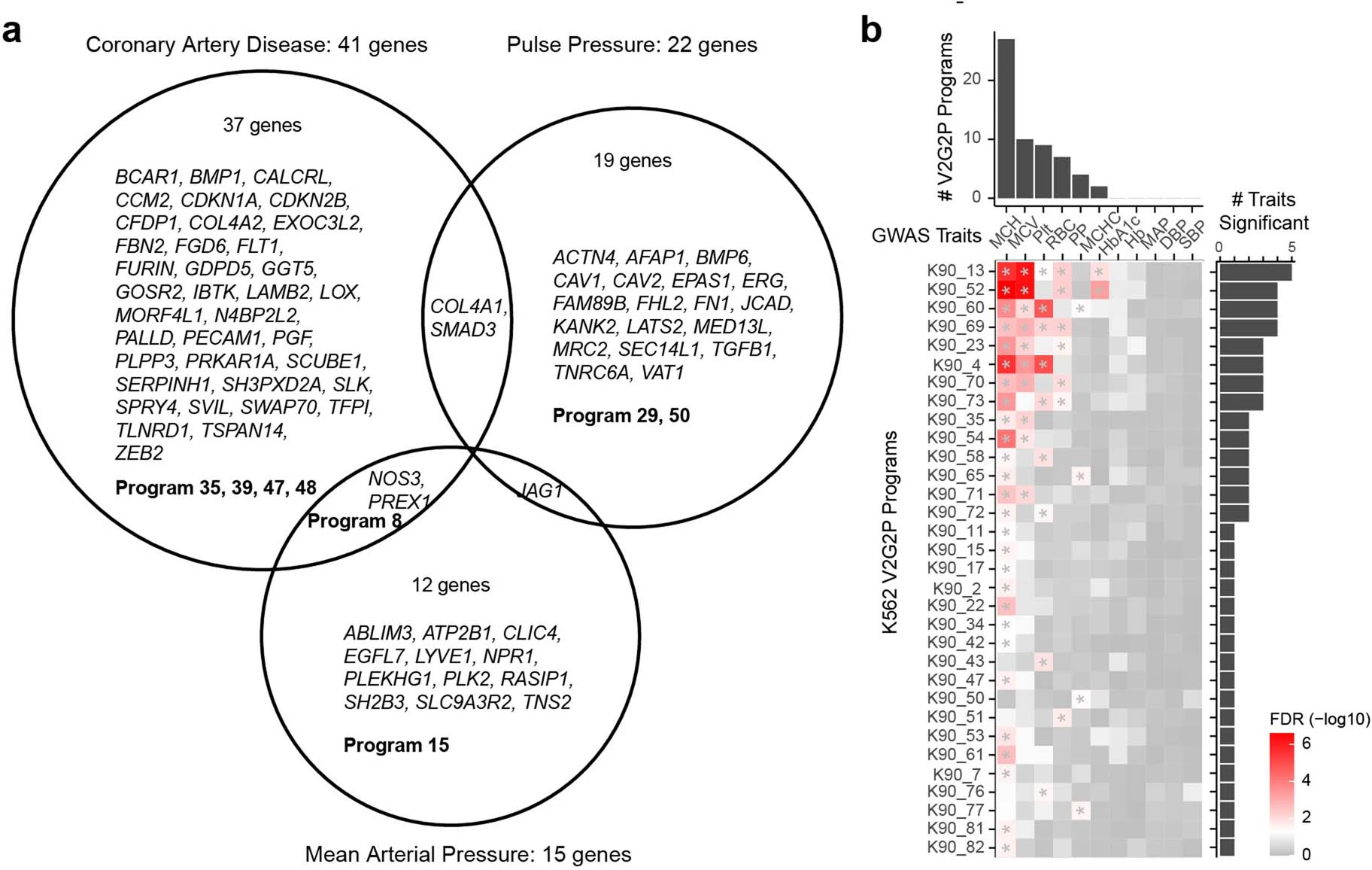
The final step involves further investigating the genes in the disease-associated programs. In the CAD study, the researchers prioritized 41 genes that may influence CAD risk through effects in endothelial cells. They performed detailed analyses on two of these genes, CCM2 and TLNRD1, demonstrating their role in regulating CAD risk genes and affecting atheroprotective processes in endothelial cells.
The V2G2P approach provides a powerful and unbiased framework for linking disease-associated variants to causal genes and pathways. By integrating diverse genomic and functional data, this method enables researchers to uncover novel disease mechanisms and potential therapeutic targets.
Application to Coronary Artery Disease
Schnitzler et al. applied the V2G2P approach to study the genetic basis of coronary artery disease (CAD), focusing on the role of endothelial cells in the development of atherosclerosis. By integrating GWAS data, epigenomic annotations, and Perturb-seq results, they identified novel insights into the molecular mechanisms underlying CAD risk.
Focusing on Endothelial Cells
The researchers began by examining the enrichment of CAD risk variants in endothelial cell enhancers. They found a significant enrichment, supporting the importance of endothelial cells in CAD pathogenesis. This finding guided their decision to perform Perturb-seq experiments in human aortic endothelial cells (teloHAECs).
Convergence of CAD Risk Loci on CCM Signaling-Related Programs
Applying the V2G2P approach, Schnitzler et al. discovered that 43 out of 306 CAD risk loci converged on five endothelial cell transcriptional programs related to the cerebral cavernous malformation (CCM) signaling pathway. This convergence suggests that the CCM pathway plays a significant role in modulating CAD risk through its effects on endothelial cell function.
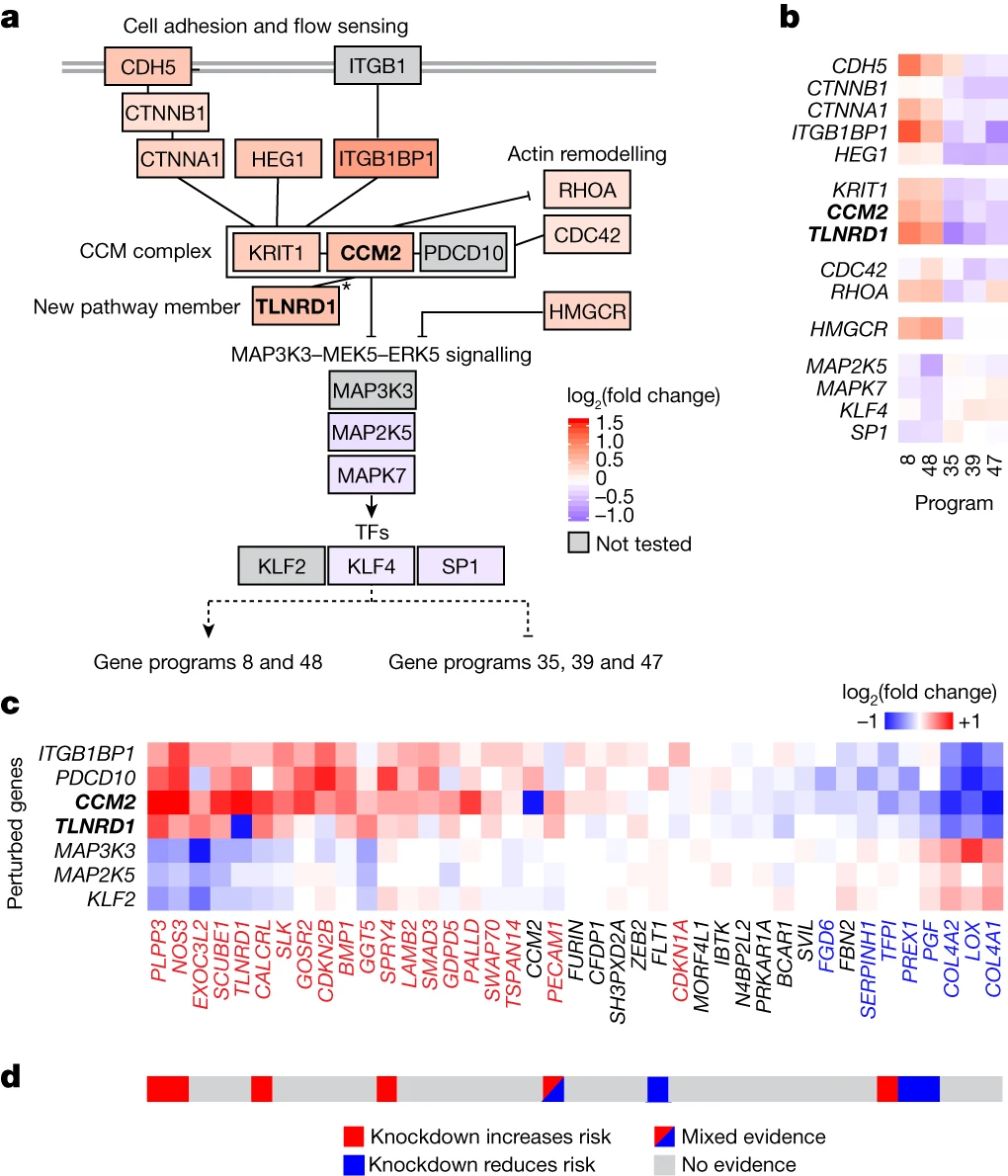
Discovering New CAD Genes: CCM2 and TLNRD1
Among the genes prioritized by the V2G2P approach, two stood out as novel regulators of the CCM pathway and CAD risk: CCM2 and TLNRD1. The researchers found that these two genes physically interact and have similar regulatory effects on the CAD-associated transcriptional programs.
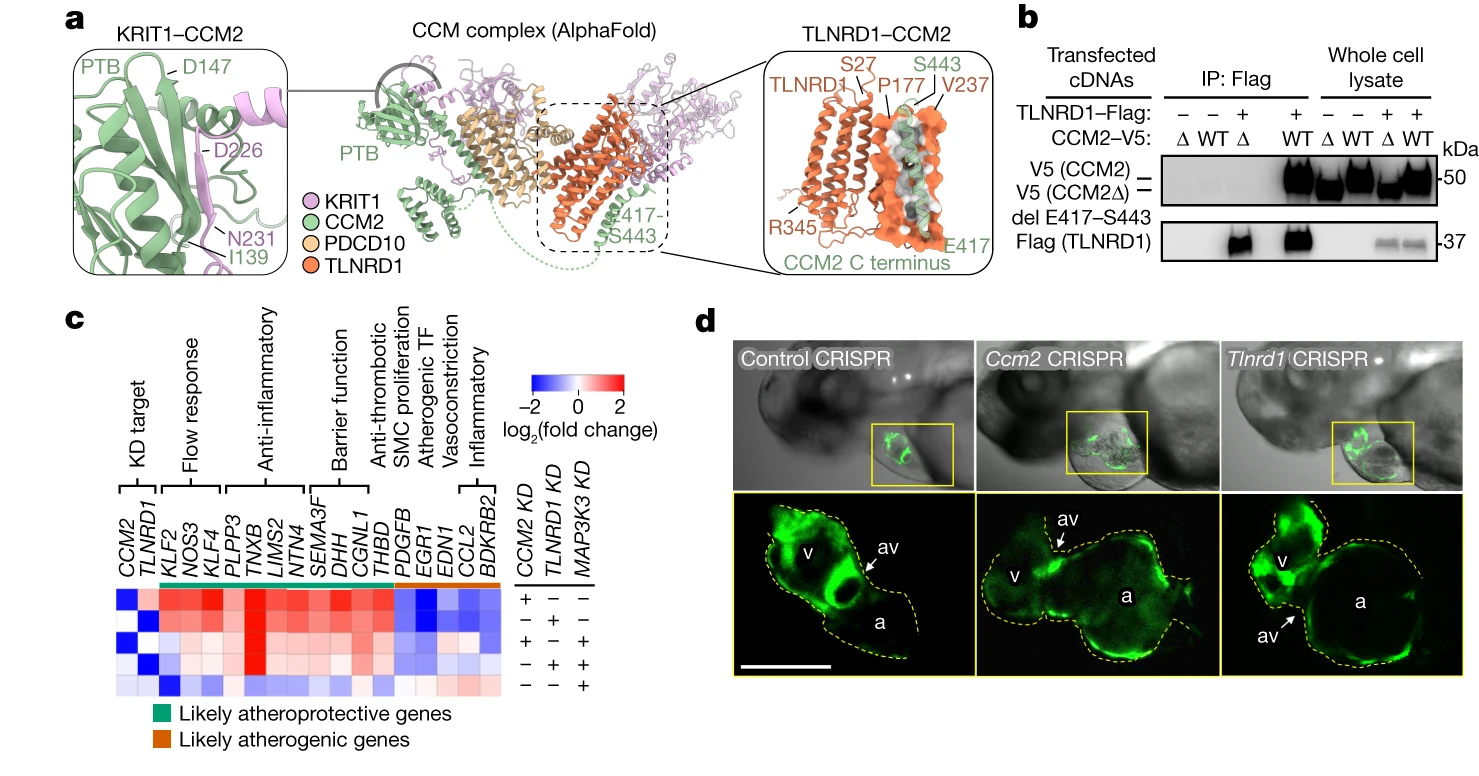
Interestingly, the effects of CCM2 and TLNRD1 knockdown on endothelial cell gene expression and phenotypes resembled those of atheroprotective laminar blood flow. This finding suggests that decreased expression of these genes, as a result of protective genetic variants, may confer resistance to atherosclerosis in regions of disturbed blood flow.
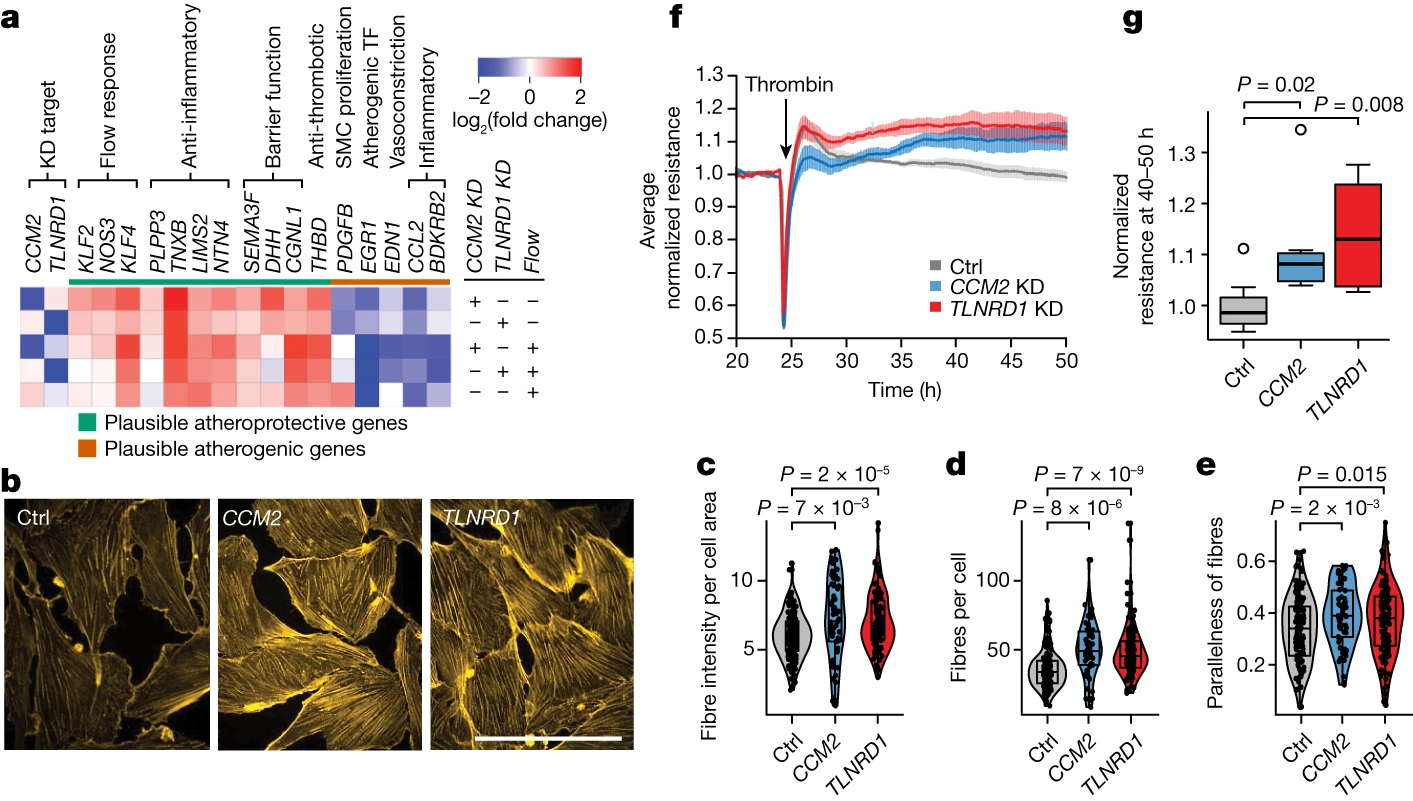
Implicating Decreased CCM Signaling as Protective in Atherosclerosis
The convergence of CAD risk loci on the CCM pathway and the atheroprotective effects of CCM2 and TLNRD1 knockdown suggest that decreased CCM signaling in endothelial cells may be protective against atherosclerosis. This finding is particularly intriguing, as it contrasts with the role of CCM signaling in the rare monogenic disease cerebral cavernous malformation, where loss-of-function mutations in CCM genes lead to vascular malformations.
The application of the V2G2P approach to CAD has uncovered a novel disease mechanism and potential therapeutic targets. By linking genetic risk factors to endothelial cell biology, this study provides a framework for understanding the complex genetic architecture of CAD and highlights the importance of cell-type-specific investigations in complex disease research.
Significance and Implications
The V2G2P approach introduced by Schnitzler et al. represents a significant advance in our ability to understand the genetic basis of complex diseases. By providing an unbiased and systematic method to link disease-associated variants to causal genes and pathways, this work has far-reaching implications for both basic research and translational medicine.
Power of an Unbiased Approach
One of the key strengths of the V2G2P approach is its ability to identify novel disease mechanisms without relying on prior knowledge of gene functions or pathways. By leveraging high-throughput single-cell perturbation screens (Perturb-seq) and unsupervised machine learning, this method can uncover previously unrecognized gene programs and regulatory relationships that are relevant to disease pathogenesis.
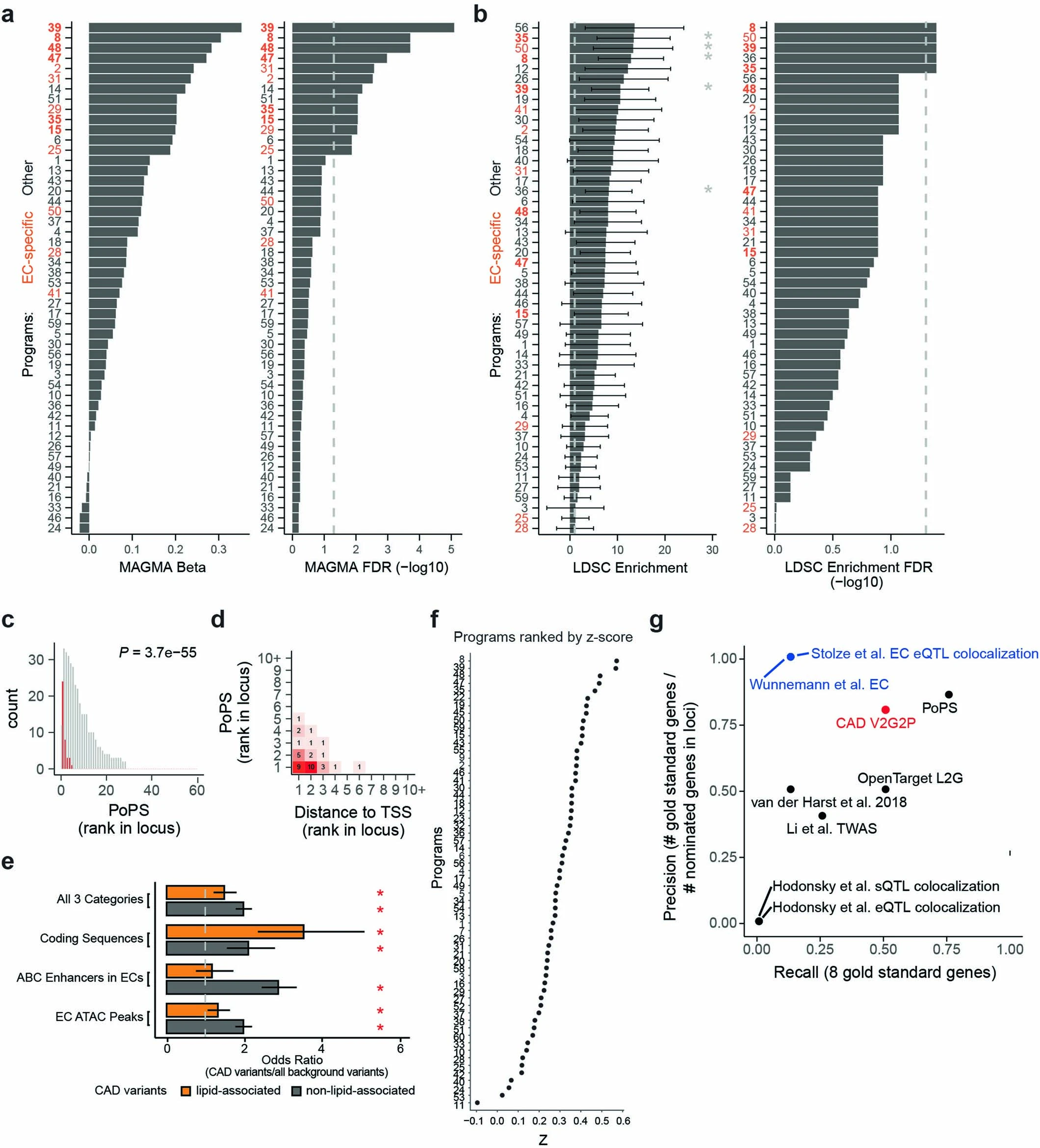
Identifying New Disease Mechanisms and Potential Therapeutic Targets
The application of V2G2P to CAD demonstrated the power of this approach to identify novel disease mechanisms and potential therapeutic targets. By uncovering the convergence of CAD risk loci on the CCM signaling pathway and the atheroprotective effects of decreased CCM signaling in endothelial cells, this study opened up new avenues for understanding and treating atherosclerosis.
Moreover, the discovery of TLNRD1 as a novel regulator of the CCM pathway and CAD risk highlights the potential of V2G2P to identify previously uncharacterized genes that play important roles in disease biology. These findings not only expand our knowledge of the molecular basis of CAD but also provide new targets for the development of therapeutic interventions.
Demonstrating the Utility of Systematic Single-Cell Perturbation Screens
The success of the V2G2P approach in elucidating the genetic basis of CAD underscores the value of systematic single-cell perturbation screens, such as Perturb-seq, in functional genomics research. By enabling the simultaneous interrogation of the effects of thousands of genetic perturbations on gene expression at single-cell resolution, these screens provide an unprecedented view of the regulatory landscape of the cell.
The integration of Perturb-seq data with GWAS results and epigenomic annotations, as demonstrated in the V2G2P approach, offers a powerful means to bridge the gap between genetic variation and cellular phenotypes. This approach can be readily extended to other cell types and complex diseases, paving the way for a more comprehensive understanding of the genetic architecture of human health and disease.
The V2G2P approach represents a significant step forward in our ability to decipher the complex genetic basis of common diseases. By providing an unbiased and systematic framework to link genetic variation to cellular pathways and disease mechanisms, this work has the potential to accelerate the discovery of novel therapeutic targets and guide the development of precision medicine strategies. As the application of this approach expands to other diseases and cell types, we can expect to gain new insights into the molecular basis of human health and disease, ultimately leading to improved diagnostic, prognostic, and therapeutic approaches.
Conclusion
The V2G2P approach introduced by Schnitzler et al. represents a major advance in our understanding of the genetic basis of complex diseases. By integrating GWAS data, epigenomic annotations, and systematic single-cell perturbation screens, this method provides an unbiased and comprehensive framework for linking disease-associated variants to causal genes and cellular pathways.
The application of V2G2P to coronary artery disease (CAD) demonstrated the power of this approach to uncover novel disease mechanisms and potential therapeutic targets. The discovery of the convergence of CAD risk loci on the CCM signaling pathway and the atheroprotective effects of decreased CCM signaling in endothelial cells highlights the importance of cell-type-specific investigations in complex disease research.
Moreover, the identification of TLNRD1 as a novel regulator of the CCM pathway and CAD risk underscores the potential of V2G2P to identify previously uncharacterized genes that play critical roles in disease biology. These findings not only expand our knowledge of the molecular basis of CAD but also provide new targets for the development of therapeutic interventions.
The success of the V2G2P approach in elucidating the genetic basis of CAD opens up exciting possibilities for its application to other complex diseases. By extending this approach to diverse cell types and disease contexts, we can expect to gain a more comprehensive understanding of the genetic architecture of human health and disease.
However, it is important to acknowledge that the application of V2G2P to other diseases and cell types may present new challenges and opportunities. Different diseases may involve distinct cell types, regulatory mechanisms, and genetic architectures, requiring adaptations and refinements of the V2G2P approach. Additionally, the availability and quality of GWAS data, epigenomic annotations, and perturbation screens may vary across diseases and cell types, necessitating careful consideration of data sources and experimental designs.
Despite these challenges, the V2G2P approach represents a significant step forward in our ability to decipher the complex genetic basis of common diseases. By providing an unbiased and systematic framework to link genetic variation to cellular pathways and disease mechanisms, this work has the potential to accelerate the discovery of novel therapeutic targets and guide the development of precision medicine strategies.
As we continue to apply and refine the V2G2P approach, we can expect to gain new insights into the molecular basis of human health and disease, ultimately leading to improved diagnostic, prognostic, and therapeutic approaches. The work of Schnitzler et al. serves as a powerful demonstration of the potential of integrating diverse genomic and functional data to unravel the genetic complexity of common diseases, paving the way for a new era of precision medicine.
This post was written with the help of Claude 3 Opus.

Leave a comment