Unraveling the Genome Evolution and Domestication History of Arabica Coffee
Summary
A groundbreaking study published in the journal Nature Genetics has provided unprecedented insights into the genome evolution and domestication history of Coffea arabica, the species responsible for the majority of global coffee production. The research team generated high-quality genome assemblies for C. arabica and its diploid progenitors, C. canephora and C. eugenioides, and performed a comprehensive analysis of their evolutionary relationships, subgenome interactions, and population genomics.
Through their analyses, the researchers uncovered key findings that shed light on the complex history of Arabica coffee, including the timing of the allopolyploid hybridization event that gave rise to the species, the patterns of gene loss and retention in its subgenomes, and the genetic basis of important agronomic traits such as disease resistance. The study also provided insights into the domestication history of Arabica coffee, revealing a clear split between wild and cultivated populations and identifying a unique genetic diversity in wild accessions from the Gesha region of Ethiopia.
The genomic resources and insights generated by this study have far-reaching implications for the future of Arabica coffee breeding and crop improvement. By providing a foundation for the development of improved molecular markers, targeted genome editing strategies, and precision breeding approaches, this research has the potential to revolutionize the coffee industry and ensure its sustainability in the face of challenges posed by climate change and disease outbreaks.
In this blog post, we will explore the key findings and implications of this groundbreaking study, delving into the details of the genomic analyses, evolutionary insights, and breeding applications that emerge from this work. Join us on a journey through the fascinating world of Arabica coffee genomics, as we unravel the secrets of this beloved crop and explore the future of coffee production in the 21st century.
Introduction
Coffea arabica, the species responsible for approximately 60% of global coffee production, is an allotetraploid hybrid of two diploid progenitor species, Coffea canephora (Robusta coffee) and Coffea eugenioides. The rich flavor and low bitterness of Arabica coffee have made it a favorite among coffee enthusiasts worldwide. However, the limited genetic diversity of cultivated C. arabica has made it susceptible to various diseases and pests, posing challenges for growers and breeders.
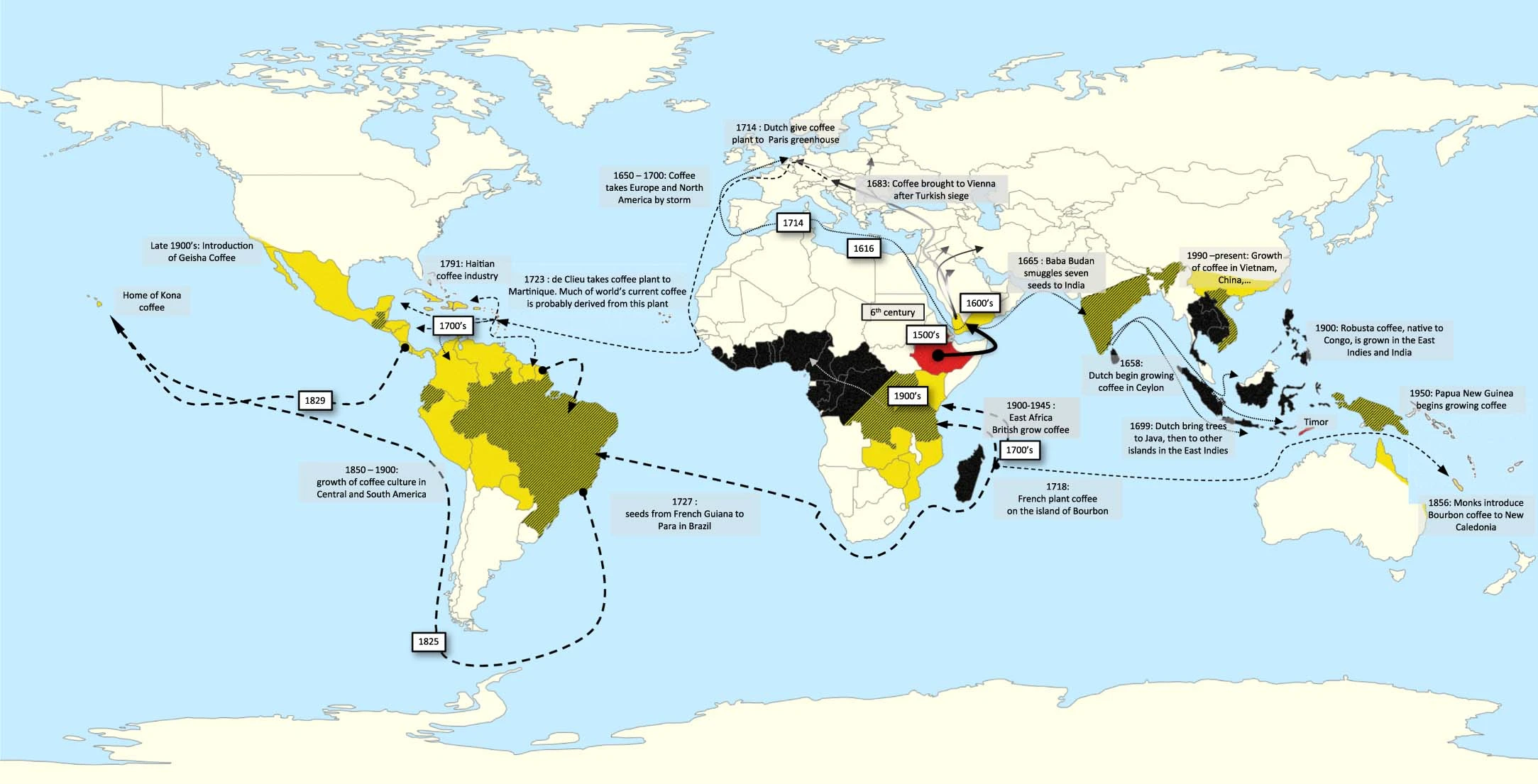
Extended Data Figure 1 from the manuscript - Coffee dissemination routes
To address these challenges and lay the foundation for future breeding efforts, a team of researchers has recently published a comprehensive study on the genome evolution and domestication history of C. arabica. In this blog post, we will delve into the key findings of this research and explore its implications for the future of Arabica coffee production.
The study, published in the journal Nature Genetics, presents high-quality, chromosome-level genome assemblies of C. arabica and its diploid progenitors, C. canephora and C. eugenioides. By comparing these genomes and analyzing the subgenomes of C. arabica, the researchers have uncovered new insights into the evolutionary history of this important crop species.
Moreover, through population genomic analysis of 41 wild and cultivated C. arabica accessions, the researchers have shed light on the domestication history of Arabica coffee, revealing the timing of key events and the relationships between wild populations and modern cultivated varieties.
Genome Sequencing and Assembly
To lay the foundation for their analysis, the researchers generated high-quality genome assemblies for C. arabica and its diploid progenitors, C. canephora and C. eugenioides. The sequencing strategies employed for each species were carefully chosen to ensure the best possible results.
For C. canephora and C. eugenioides, the researchers used a combination of long-read PacBio sequencing and short-read Illumina sequencing, followed by Hi-C scaffolding. This approach allowed them to generate chromosome-level assemblies for both species, with sizes of 672 Mb and 645 Mb, respectively.
| Assembly | C. eugenioides | C. canephora | C. arabica | C. arabica HiFi |
|---|---|---|---|---|
| Projected genome size (Mb)a | 682 | 705 | 1,281 | 1,281 |
| Total assembly length (Mb) | 661 | 672 | 1,088 | 1,198 |
| % of projected genome | 96.90% | 95.30% | 84.90% | 93.50% |
| N scaffolds | 253 | 3,033 | 8,474 | 132 |
| Scaffold N50 | 61.3 Mb | 50.1 Mb | 32.7 Mb | 53.7 Mb |
| N contigs | 5,736 | 3,757 | 11,863 | 238b |
| Contig N50c (Mb) | 0.4 | 1.35 | 0.23 | 30 |
| Pseudochromosomes (Mb) | NAd | 583 | 801 | 1,192 |
| % of projected genome | NA | 82.70% | 62.50% | 93.10% |
| N genes | 32,192 | 28,880 | 56,670 | 69,314 |
| Genes in pseudochromosomes | NA | 27,881 | 50,410 | 69,067 |
| % genes in pseudochromosomes | NA | 97% | 89% | 99.60% |
| BUSCO genome | ||||
| Complete | 96.70% | 97.40% | 97.60% | 97.90% |
| Single | 88.50% | 94.80% | 20.10% | 4.30% |
| Duplicated | 8.20% | 2.60% | 77.50% | 93.60% |
| Fragmented | 1.10% | 0.90% | 0.80% | 0.80% |
| Missing | 2.20% | 1.70% | 1.60% | 1.30% |
| Total | 2,326 | 2,326 | 2,326 | 2,326 |
| BUSCO annotation | ||||
| Complete | 94.90% | 96.20% | 92.10% | 97.30% |
| Single | 82.40% | 92.80% | 33.30% | 4.10% |
| Duplicated | 12.50% | 3.40% | 58.80% | 93.20% |
| Fragmented | 2.10% | 1.50% | 2.80% | 0.80% |
| Missing | 3.00% | 2.30% | 5.10% | 1.90% |
| Total | 2,326 | 2,326 | 2,326 | 2,326 |
Table 1 from the manuscript - Statistics of the Coffea assemblies presented in this paper
The C. arabica genome, being an allotetraploid, required a more complex approach. The researchers sequenced a di-haploid C. arabica accession using a combination of PacBio and Illumina sequencing. They then used Hi-C scaffolding to generate a chromosome-level assembly, which spanned 1,088 Mb. To further improve the C. arabica assembly, they generated a second assembly using PacBio HiFi long-read sequencing followed by Hi-C scaffolding. This high-quality assembly spanned 1,198 Mb, with 1,192 Mb anchored to pseudochromosomes.
The completeness of these genome assemblies was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCOs), which revealed that all assemblies had a completeness of >96%. Notably, the C. arabica HiFi assembly showed that 93.2% of the BUSCO genes were duplicated, indicating that most gene duplicates from the allopolyploidy event were retained.
These high-quality genome assemblies serve as the backbone for the subsequent analyses of genome evolution, subgenome interactions, and population genomics in C. arabica. The availability of chromosome-level assemblies for both the allotetraploid C. arabica and its diploid progenitors enables a detailed comparison of their genomes, shedding light on the evolutionary processes that have shaped the Arabica coffee genome.
Subgenome Analysis and Evolution
One of the key aspects of this study was the analysis of the two subgenomes within C. arabica, derived from its diploid progenitors C. canephora (subCC) and C. eugenioides (subEE). By comparing these subgenomes to each other and their diploid counterparts, the researchers aimed to uncover the evolutionary processes that have shaped the C. arabica genome since the allopolyploid hybridization event.
Remarkably, the researchers found that the two subgenomes of C. arabica have remained highly conserved, with no evidence of major “genomic shock” after hybridization. This is in contrast to some other allopolyploid species, where hybridization has been followed by significant genomic rearrangements and changes in transposable element activity.
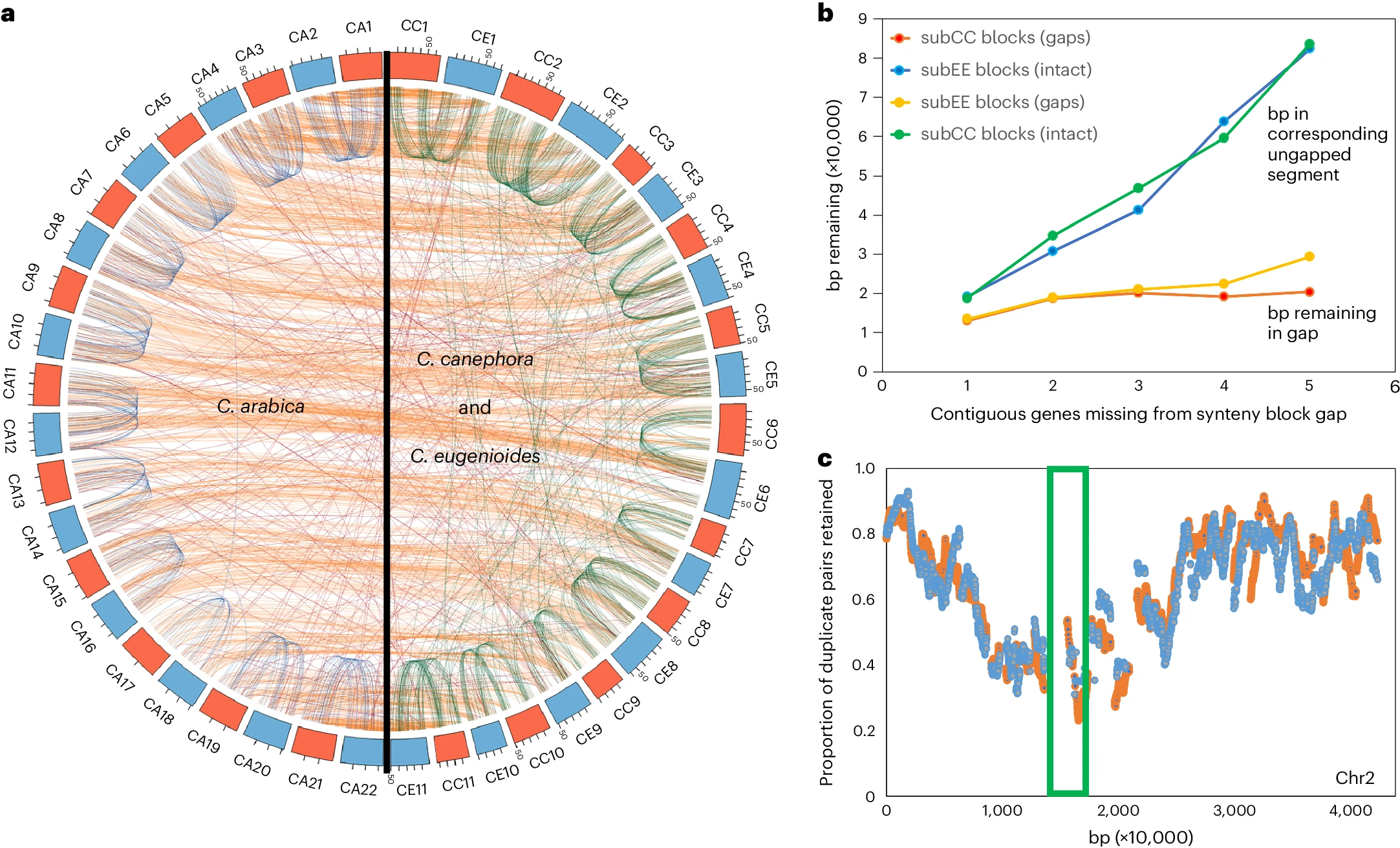
Figure 1 from the manuscript - Patterns of synteny, fractionation and gene loss in C. arabica and its progenitor species C. canephora and C. eugenioides
The researchers also investigated the patterns of gene loss and genome fractionation in the subgenomes of C. arabica and its progenitors. They found that the rates of genome fractionation were similar before and after the hybridization event, suggesting that the allopolyploidy did not significantly accelerate or slow down the process of gene loss.
Interestingly, the study revealed that the main mechanism of genome fractionation in these species has been through small-scale deletions of one or a few genes at a time, rather than large-scale deletions or rearrangements. These deletions were found to occur more frequently in pericentromeric regions of the chromosomes, while the chromosome arms showed more moderate levels of gene loss.
Furthermore, the researchers found evidence for biased fractionation, with genes involved in certain biological processes being more likely to be retained in duplicated copies, while others were more prone to loss. This pattern is consistent with the gene dosage balance hypothesis, which suggests that genes involved in complex cellular processes and networks are more likely to be retained in duplicate after polyploidy events.
These findings provide valuable insights into the evolutionary dynamics of allopolyploid genomes and the mechanisms that shape their structure and function over time. By understanding these processes in C. arabica, researchers can better predict how the genome may continue to evolve in the future and how this may impact important agronomic traits.
Gene Expression and Subgenome Dominance
In allopolyploid species, the presence of two or more distinct subgenomes can lead to complex patterns of gene expression and regulation. In some cases, one subgenome may dominate over the other(s), a phenomenon known as subgenome dominance. To investigate whether this occurs in C. arabica, the researchers analyzed gene expression patterns across the two subgenomes.
Surprisingly, the study found no evidence of global subgenome dominance in C. arabica. In other words, neither the C. canephora-derived subgenome (subCC) nor the C. eugenioides-derived subgenome (subEE) showed consistently higher or lower levels of gene expression across all tissues and developmental stages.
However, when the researchers looked at specific gene families, they did find evidence of localized differences in expression between the two subgenomes. For example, in the gene families encoding enzymes involved in the biosynthesis of caffeine, terpenoids, and fatty acids (all important components of coffee bean chemistry and flavor), some genes showed higher expression levels in one subgenome than the other.
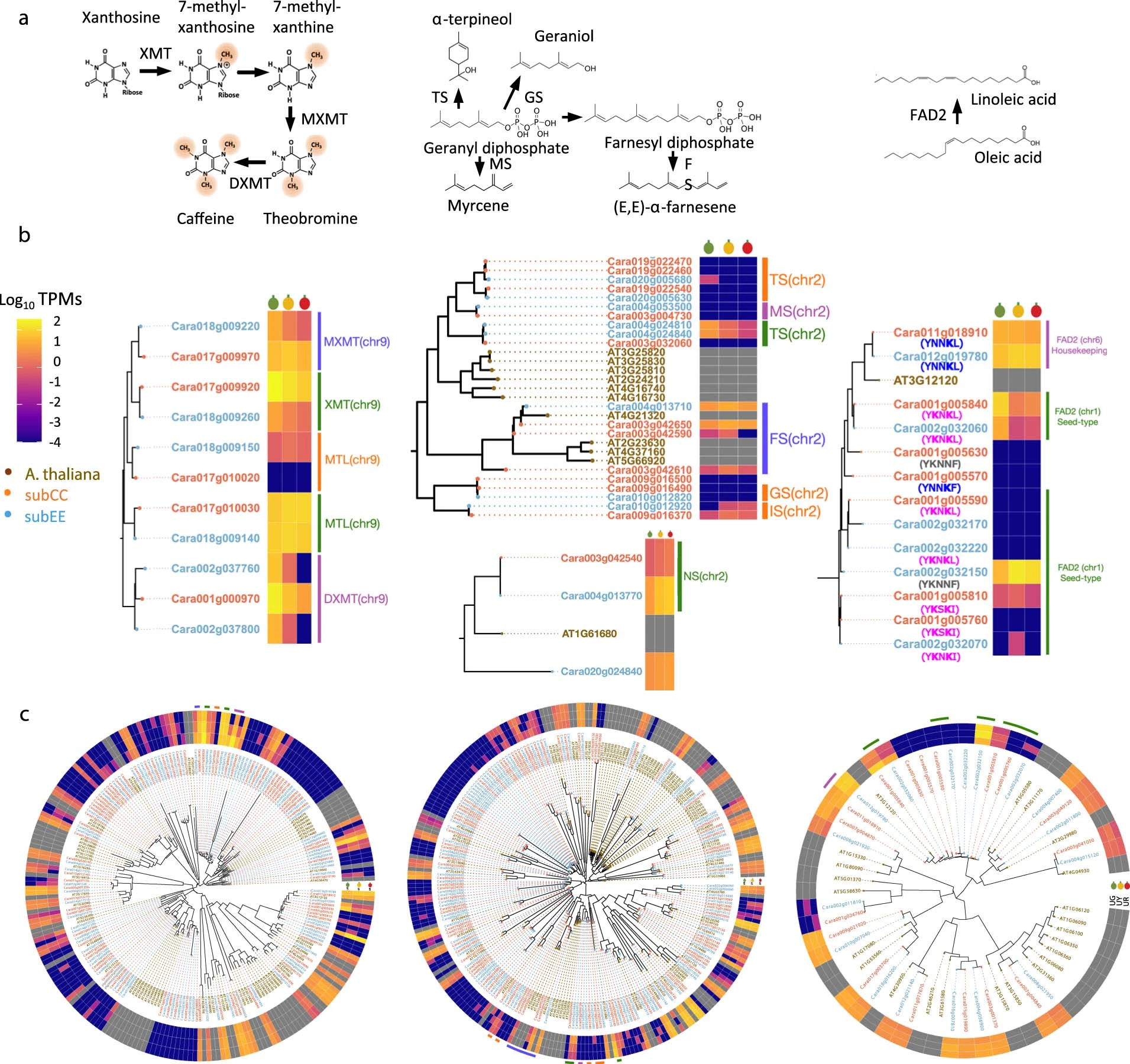
Extended Data Figure 2 from the manuscript - Composition and expression of exemplar Arabica gene families contributing to bean quality traits
These findings suggest that while there is no global subgenome dominance in C. arabica, the two subgenomes may have evolved to specialize in the expression of certain gene sets. This mosaic pattern of subgenome expression has been observed in other allopolyploid species, such as rapeseed and cotton, and is thought to be a common feature of recently formed allopolyploids.
The lack of global subgenome dominance in C. arabica may be due to the relatively recent origin of this species (estimated to have occurred between 350,000 and 610,000 years ago) and the high degree of similarity between its progenitor species. As the subgenomes continue to evolve and differentiate over time, it is possible that more pronounced patterns of subgenome dominance may emerge.
These findings have important implications for understanding the genetic basis of key agronomic traits in C. arabica, such as flavor profile and disease resistance. By identifying the specific genes and subgenomes that contribute to these traits, researchers can develop more targeted breeding strategies to improve the quality and resilience of Arabica coffee.
Population Genomics and Domestication History
To gain insights into the evolutionary history and domestication of C. arabica, the researchers sequenced and analyzed the genomes of 41 wild and cultivated accessions from various locations and breeding programs. This population genomic analysis revealed several key findings that shed light on the origin and spread of Arabica coffee.
First, the researchers estimated that the allopolyploid hybridization event that gave rise to C. arabica occurred between 350,000 and 610,000 years ago. This is earlier than some previous estimates, which had suggested a more recent origin. Following this hybridization event, the species underwent several population bottlenecks, leading to a reduction in genetic diversity.
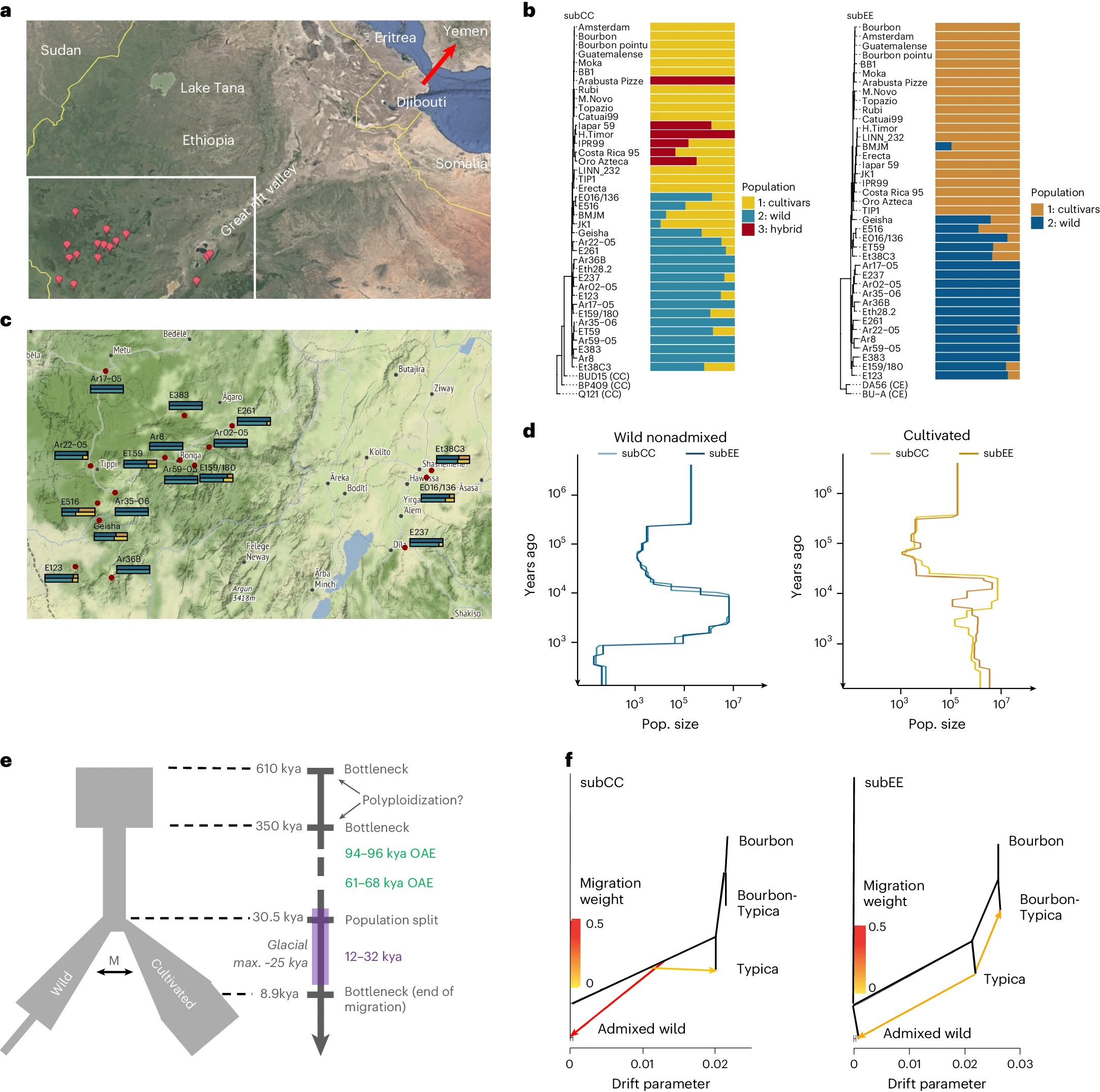
Figure 2 from the manuscript - Population history of C. arabica
By comparing the genomes of wild and cultivated accessions, the researchers were able to identify a clear split between these two groups, which occurred around 30,500 years ago. This suggests that the domestication of C. arabica began relatively early in its evolutionary history. Interestingly, the study also found evidence of ongoing gene flow between wild and cultivated populations until around 8,900 years ago, indicating that the process of domestication was gradual and involved continued exchange with wild populations.
The researchers also investigated the genetic relationships between different cultivated varieties of C. arabica. They found that the two main cultivar groups, Typica and Bourbon, are closely related and share a common origin, likely tracing back to a small number of plants that were transported out of Ethiopia in the 16th and 17th centuries. These findings highlight the narrow genetic base of cultivated Arabica coffee and the challenges this poses for breeding and crop improvement.
Interestingly, the study also identified a group of wild accessions from the Gesha region of Ethiopia that are genetically distinct from other wild populations and show close affinity to the cultivated varieties. This suggests that the Gesha region may have been an important center of origin for the domestication of C. arabica, and that the unique genetic diversity found in this region could be valuable for future breeding efforts.
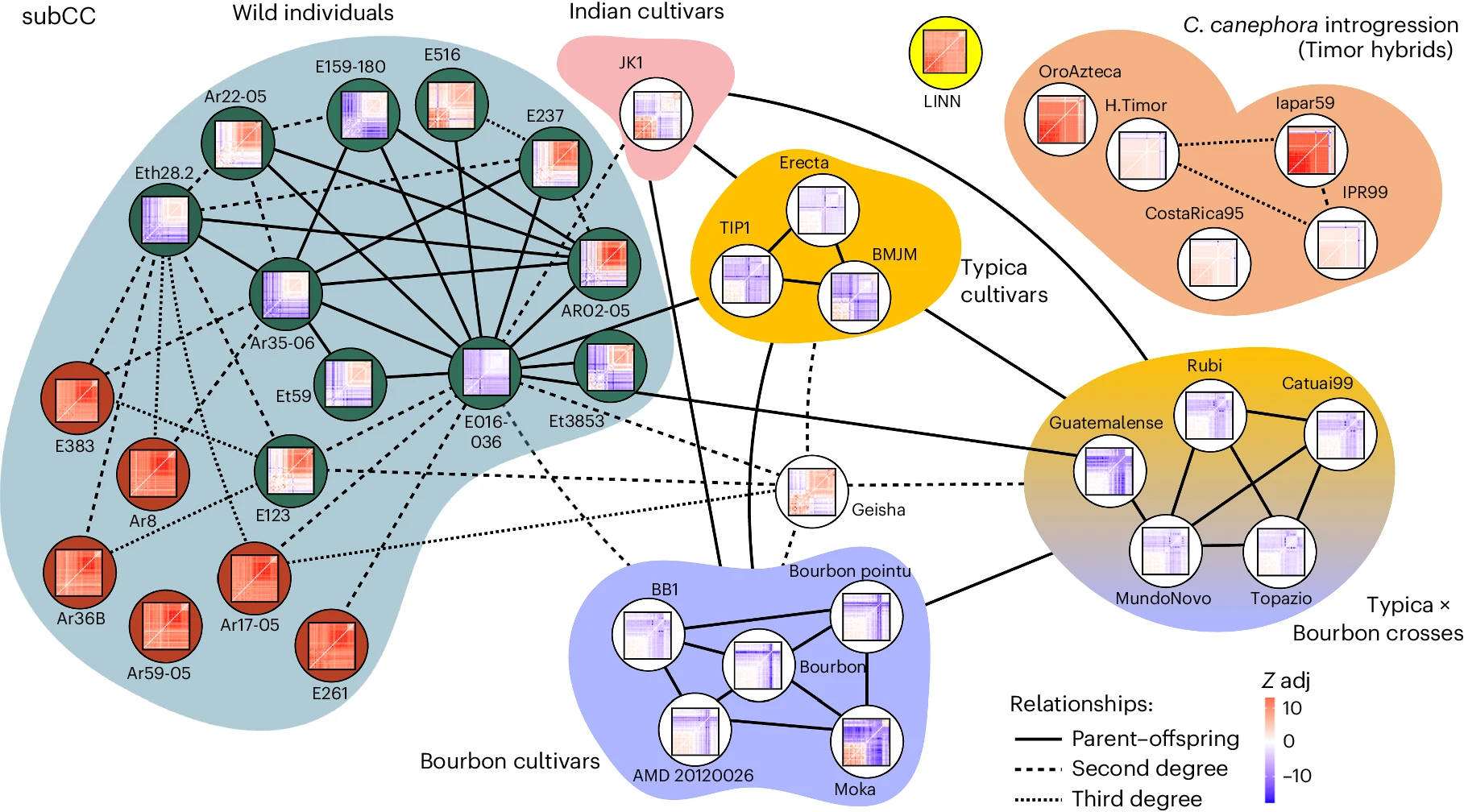
Figure 3 from the manuscript - Kinship estimation of C. arabica accessions, inferred from SNPs in subCC
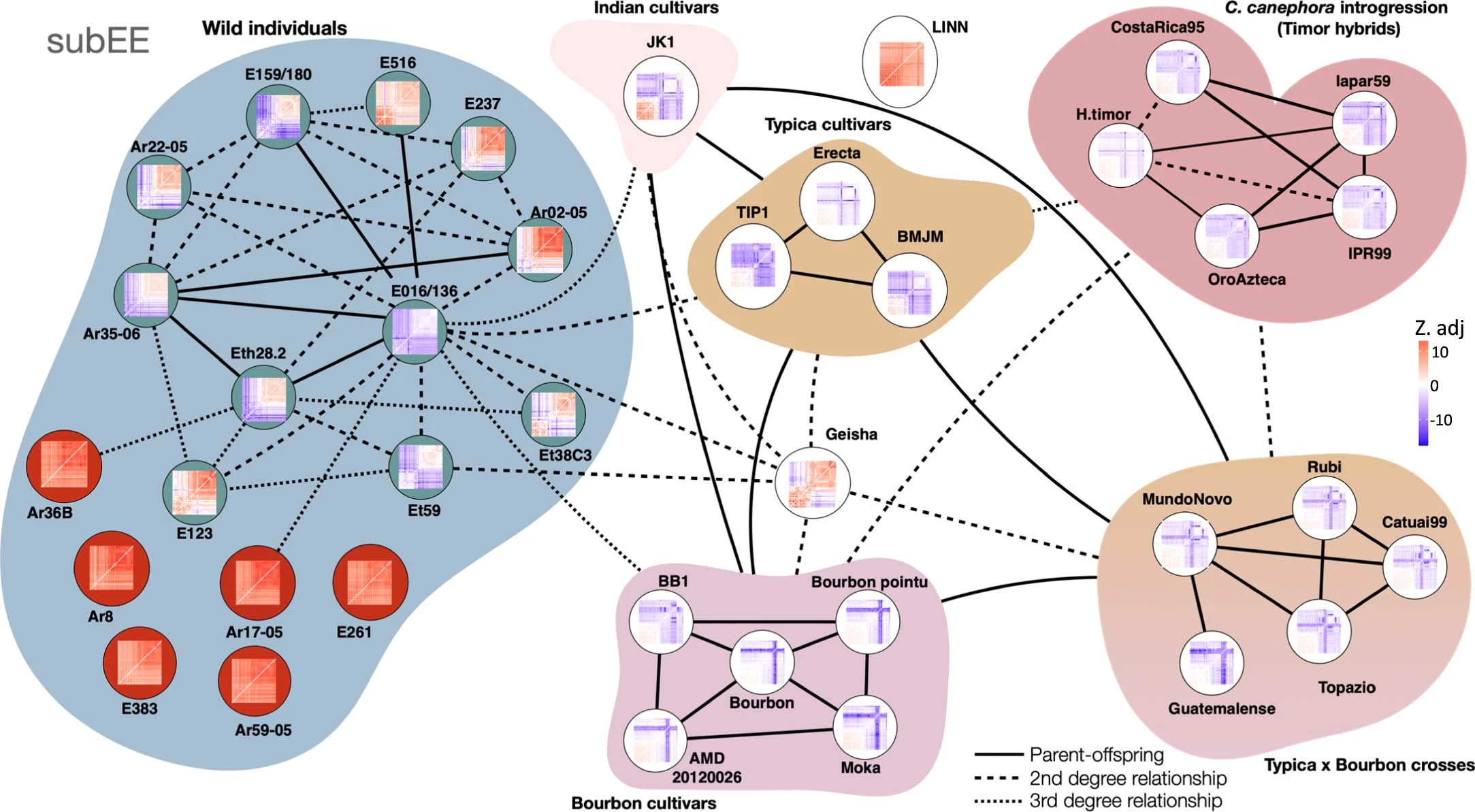
Extended Data Figure 7 from the manuscript - Kinship analysis on SubEE
Finally, the researchers investigated the genetic basis of disease resistance in C. arabica by analyzing a group of cultivars that are derived from a spontaneous hybrid between C. canephora and C. arabica, known as the Timor hybrid. They found that these cultivars have inherited large segments of the C. canephora genome, particularly in regions that contain disease resistance genes. This introgression of genetic material from C. canephora has been an important source of disease resistance in modern Arabica cultivars.
Overall, these findings provide a detailed picture of the evolutionary history and domestication of C. arabica, and highlight the challenges and opportunities for improving this important crop through breeding and genetic analysis.
Introgression and Disease Resistance
One of the major challenges facing Arabica coffee production is the crop’s susceptibility to various diseases, particularly coffee leaf rust caused by the fungal pathogen Hemileia vastatrix. To combat this problem, breeders have often turned to introgression, the transfer of genetic material from one species to another through hybridization and backcrossing. In C. arabica, a key source of disease resistance has been the spontaneous hybrid between C. canephora and C. arabica, known as the Timor hybrid.
In this study, the researchers investigated the genomic basis of disease resistance in C. arabica by analyzing a group of cultivars derived from the Timor hybrid. They found that these cultivars have inherited large segments of the C. canephora genome, particularly in regions that contain disease resistance genes.
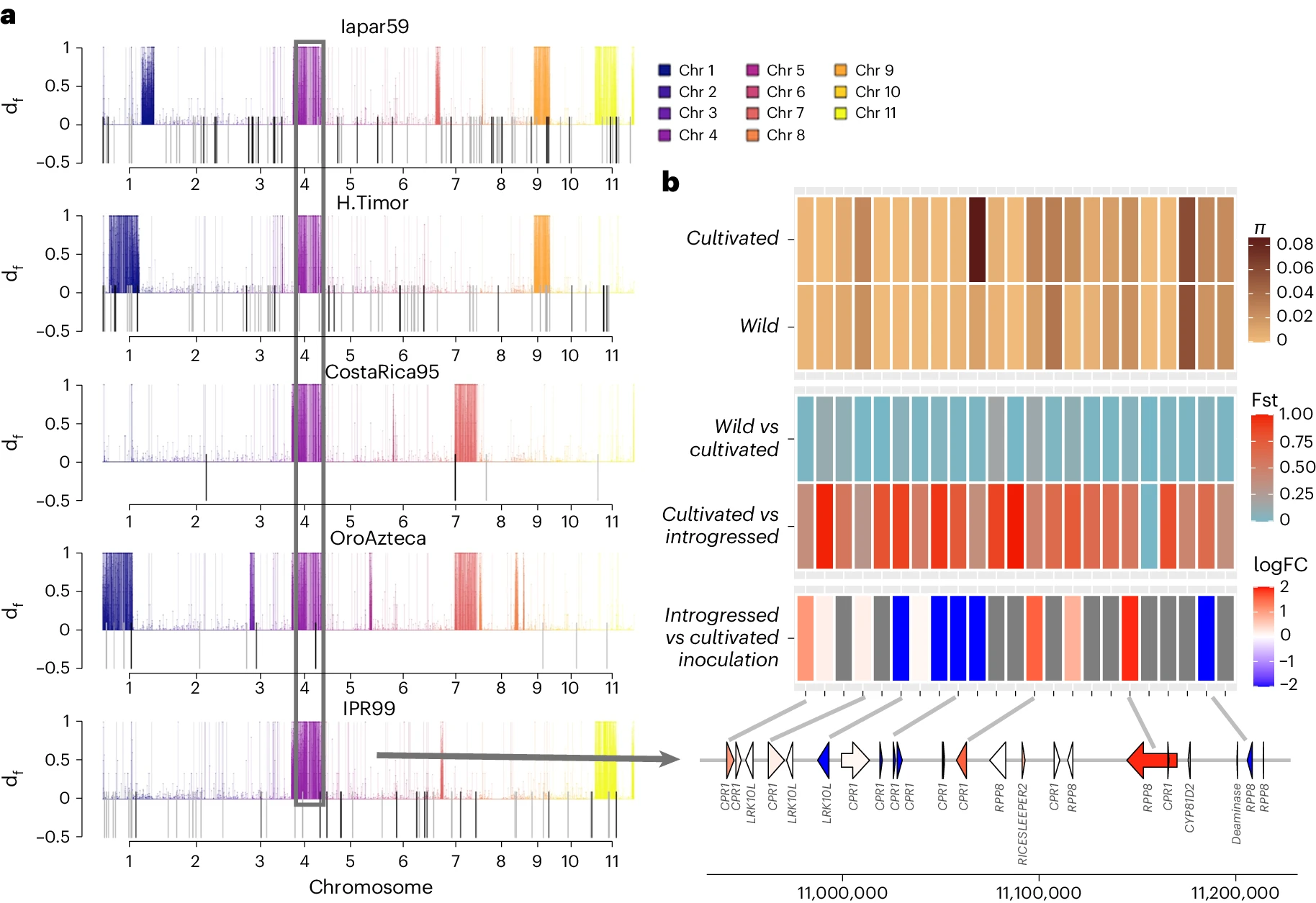
Figure 4 from the manuscript - Introgression of C. canephora into H. vastatrix-resistant C. arabica lineages
By comparing the genomes of the Timor hybrid-derived cultivars to those of other C. arabica accessions, the researchers were able to identify specific regions of the genome that have undergone introgression from C. canephora. These regions were found to be enriched in genes associated with disease resistance, including nucleotide-binding leucine-rich repeat (NLR) genes, which are known to play a key role in plant immune responses.
Interestingly, the study also found that the introgressed regions from C. canephora tend to have higher levels of genetic diversity than the surrounding C. arabica genome. This suggests that the introgression of genetic material from C. canephora has not only provided disease resistance genes but has also increased the overall genetic diversity of the Timor hybrid-derived cultivars.
Further analysis of gene expression patterns in the introgressed regions revealed that several disease resistance genes show higher levels of expression in the Timor hybrid-derived cultivars compared to susceptible C. arabica accessions. This includes genes encoding proteins involved in pathogen recognition, signaling, and defense responses.
These findings highlight the importance of introgression as a tool for improving disease resistance in C. arabica and provide valuable insights into the specific genes and genomic regions that contribute to this important trait. By understanding the genetic basis of disease resistance, breeders can develop more targeted strategies for introducing resistance genes into elite Arabica cultivars while minimizing the impact on other desirable traits such as flavor and aroma.
Moreover, the identification of disease resistance genes in the Timor hybrid-derived cultivars opens up new opportunities for using modern breeding technologies, such as marker-assisted selection and genome editing, to accelerate the development of disease-resistant Arabica varieties. As climate change and the spread of pathogens continue to pose challenges for coffee production, the insights gained from this study will be invaluable for ensuring the sustainability and resilience of the Arabica coffee industry.
Future Perspectives and Breeding Applications
The findings presented in this study have far-reaching implications for the future of Arabica coffee breeding and crop improvement. By providing high-quality genome sequences, detailed analyses of subgenome evolution and interaction, and insights into the domestication history and disease resistance of C. arabica, this research lays the foundation for a new era of precision breeding in coffee.
One of the most immediate applications of this work will be in the development of improved molecular markers for key agronomic traits in Arabica coffee. The identification of specific genes and genomic regions associated with disease resistance, flavor, and other important characteristics will enable breeders to design targeted markers for use in marker-assisted selection. This will allow for more efficient and precise breeding strategies, accelerating the development of new cultivars with improved performance and resilience.
Another exciting possibility opened up by this research is the use of genome editing technologies, such as CRISPR-Cas9, to introduce targeted modifications into the Arabica coffee genome. With the availability of high-quality reference genomes and a better understanding of the genetic basis of key traits, researchers can now design precise genome editing strategies to enhance specific characteristics while minimizing off-target effects. This could lead to the development of new Arabica varieties with improved disease resistance, enhanced flavor profiles, or increased tolerance to environmental stresses.
The insights gained from the population genomic analysis of wild and cultivated Arabica accessions will also be valuable for guiding future germplasm collection and conservation efforts. The identification of unique genetic diversity in wild populations, particularly those from the Gesha region of Ethiopia, highlights the importance of preserving and studying these resources for potential use in breeding programs. By incorporating this diversity into cultivated backgrounds, breeders may be able to expand the genetic base of Arabica coffee and develop new varieties with improved resilience and adaptability to changing climatic conditions.
Finally, the detailed analysis of subgenome evolution and interaction in C. arabica provides a model for studying the dynamics of allopolyploid evolution in other important crop species. Many major crops, including wheat, cotton, and rapeseed, are also allopolyploids, and understanding how their subgenomes evolve and interact over time is crucial for developing effective breeding and crop improvement strategies. The approaches and insights gained from this study can be applied to other allopolyploid crops, advancing our understanding of genome evolution and its implications for agriculture.
In conclusion, the groundbreaking research presented in this study represents a major milestone in our understanding of the complex evolutionary history and genetics of Arabica coffee. By providing a wealth of genomic resources and insights, this work opens up new avenues for breeding and crop improvement, with the potential to revolutionize the future of the coffee industry. As we continue to face challenges posed by climate change, disease outbreaks, and increasing demand for high-quality, sustainable coffee, the knowledge gained from this study will be invaluable for ensuring the long-term success and resilience of Arabica coffee production.
This post was written with the help of Claude 3 Opus.

Leave a comment