[Paper review] Sequence modeling and design from molecular to genome scale with Evo
Introduction
DNA language model
- DNA as the fundamental layer of biological information –> DNA sequencing enabled the systematic mapping of the evolutionary diversity at the the whole-genome scale
- Towards a general biologiccal foundation model that learns the intrinsic logic of whole genome current efforts to model molecular biology with ML: modality-specific (proteins, regulatory DNA, RNA), design of single molecule, simple complexes, short DNA sequences
- A DNA model that unifies information across the molecular, systems and genome scale could learn systems-wide interactions, enable the design of more sophisticated biological functions
LLM in biology
-
recent success of LLM using transformer architecture
- existing attempts to model DNA as a language: limited by computational cost, generally underperforms at single-nucleotide or byte-level resolution
- transformer-based DNA models: constrained to short context, sacrifice single-nucleotide resolution by aggregating nucleotides to tokens
-
Evo: a 7B genomic foundation model, trained to generate DNA sequences at whole-genome scale
- context length of 131k tokens
- based on StripedHyena architeccture: hybridizes attention and data-controlled convolutional operators, efficiently deal with long sequences
- trained on prokaryotic whole-genome dataset (300B nucleotides)
- byte-level, single-nucleotide tokenizer
Introduction to Evo
- Downstream tasks – used in both prediction and generation tasks at the molecular, systems, and genome scale
- zero-shot prediction
- predicting the fitness effects of mutations on proteins
- predicting the fitness effects of mutations on noncoding RNAs
- predicts the combinations of prokaryotic promoter-ribosome binding site (RBS) pairs from regulatory sequence alone
- designing synthetic multi-component biological systems
- learns the co-evolutionary linkage of coding & noncodoing sequences
- designed CRISPR-Cas systems, and transposable elements
- whole-genome scale
- can predict essential genes in bacteria/bacteriophages without any supervision
- generated sequesnces over 650kb with plausible genomic coding architecture
- zero-shot prediction
-> Evo establishes a foundational paradigm for predictive & generative biological sequence modeling
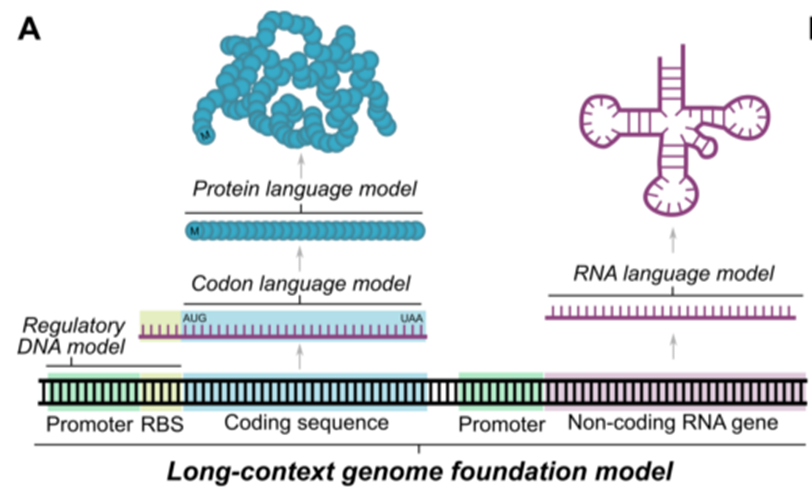
Model architecture
StripedHyena architecture
- first alternative model architecture competitive with ‘Transformers’
- efficiet autoregressive generation
- low latency, faster decoding, higher throughput than transformers
- faster training & finetuning at long context (>3x at 131k)
- robust to training beyond the compute-optimal frontier
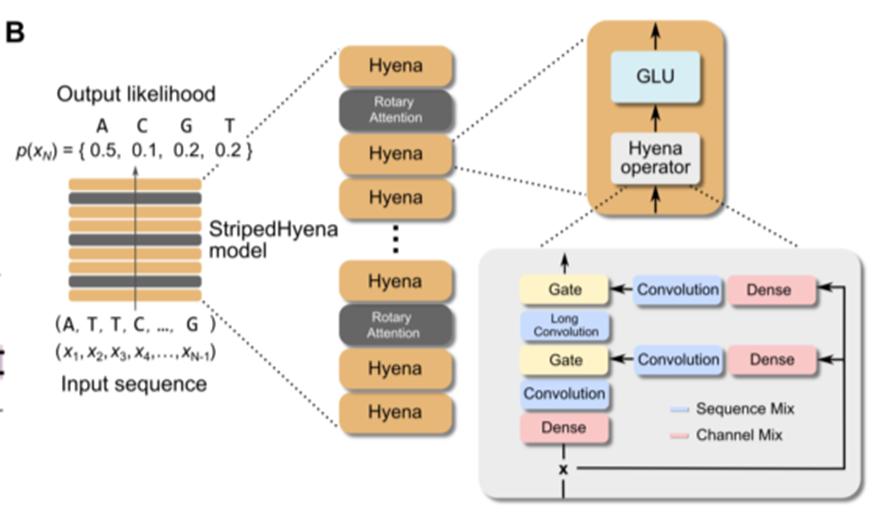
- hybrid of 29 layers of Hyena layers (data-controlled convolutional operators)
-
interleaved with 3 layers (10%) of multi-head attention equipped with RoPE (rotary position embeddings)
- Hyena Hierarchy: Towards Larger Convolutional Language Models
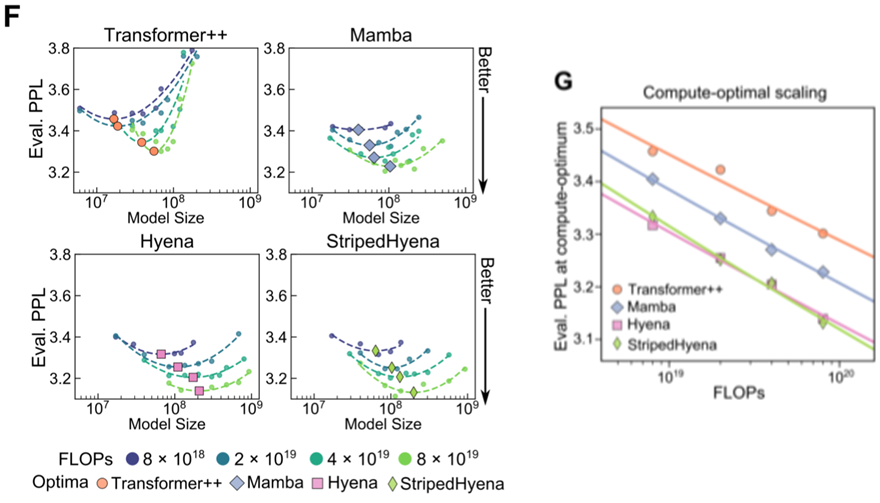
- StripedHyena was an optimal architecture for long DNA sequences pretraining
- Compared to the previous DNA model HyenaDNA, which also utilizes the Hyena architecture, the model size has expanded by over 1000 times and the data by over 100 times.
- maximum number of tokens (base count) at compute-optimal (minimum Eval. perplexity): 250 billion
Training data
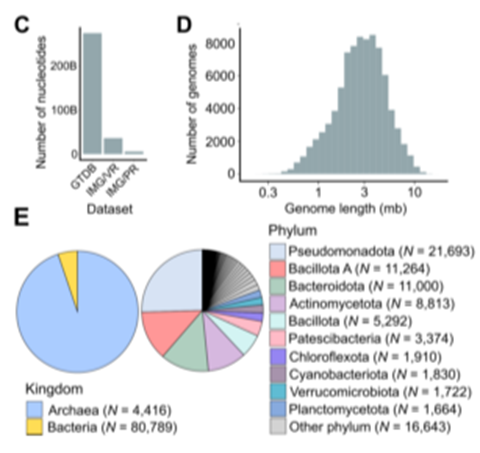
- trained with GTDB, IMG/PR and viral sequences from IMG/VR
- viruses that infect eukaryotic hosts were excluded
Zero-shot function prediction
Predicting mutational effects on protein function
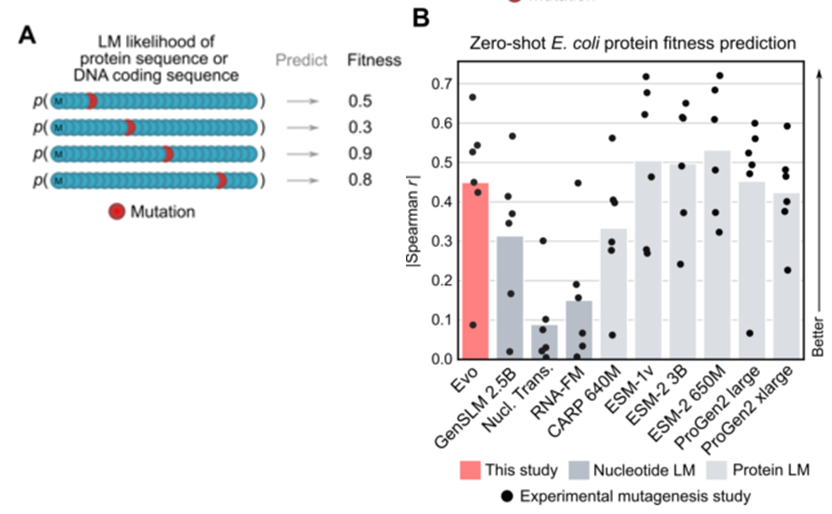
- Predicting fitness (study-specific metric quantifying how well a protein performs a certain function) upon mutation
-
Utilizing deep mutational scanning (DMS) dataset -> exhaustive set of mutations to a protein coding sequence, experimentally measured fitness
- Competitive performance despite not learning protein language, only using DNA
- However, failed to predict mutational effects on human proteins DMS dataset as it was pre-trained only on prokaryote data
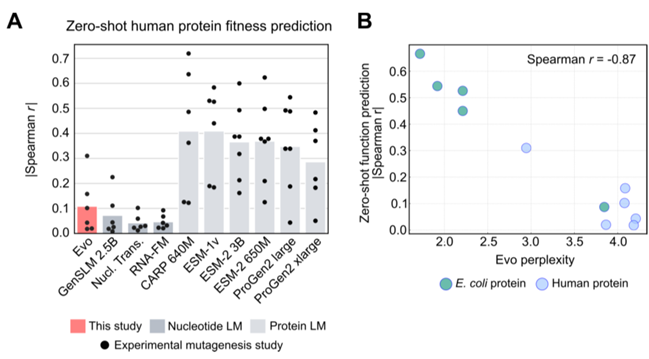
Predicting mutational effects on ncRNA function / predicting gene expression from regulatory DNA
Fine-tuning: Generative design of coplex systems
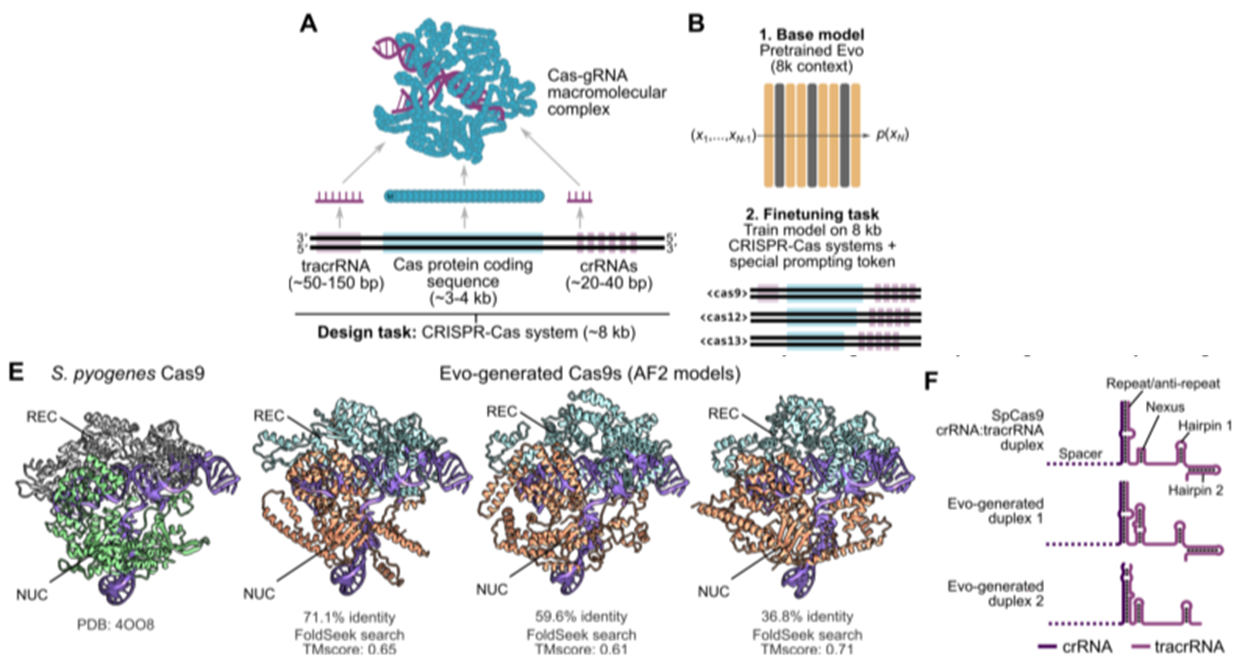
Generative design of CRISPR-Cas molecular complexes
- Finetuning on generating CRISPR-Cas system (~8kb)
Generative design of transposable biological systems
- finetuning on generating IS200/IS605 system (~2kb)
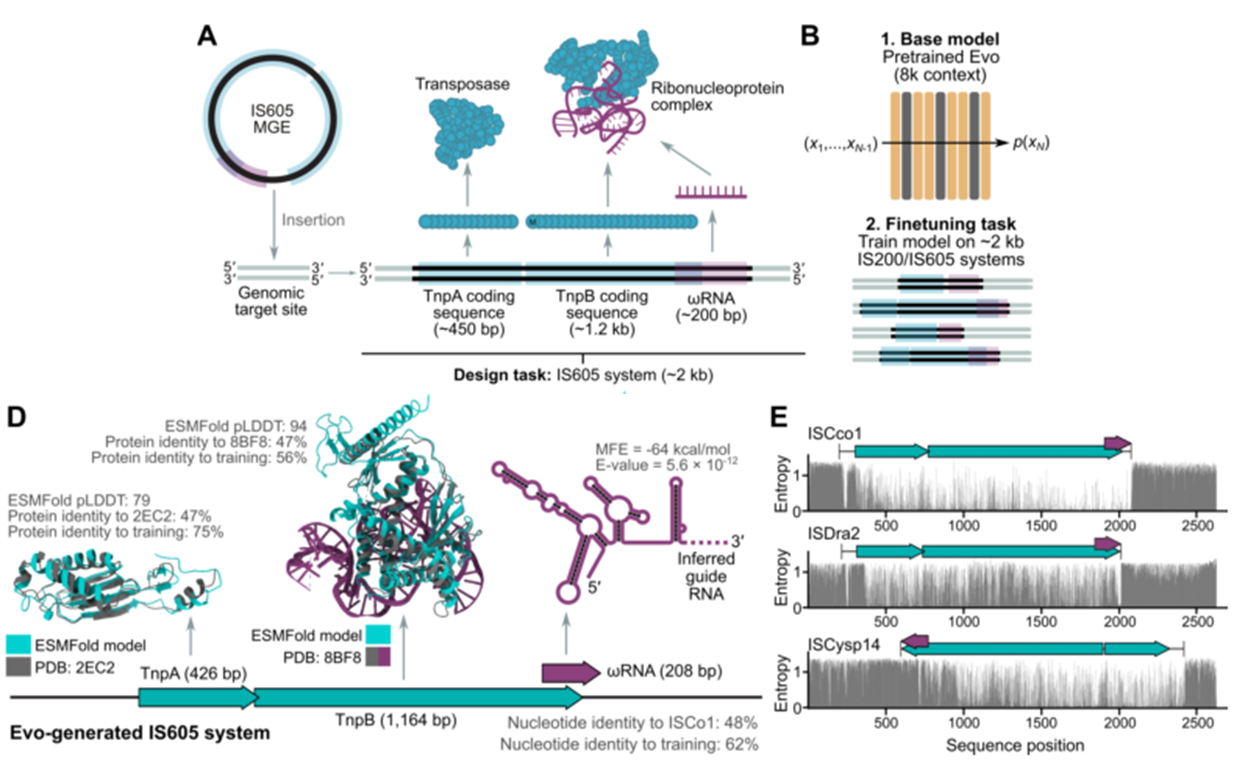
Analyzing whole genome
Predicting gene essentiality with long genomic context
- second-stage pretraining with species-level special tokens: extending context to 131k-long genomic segments
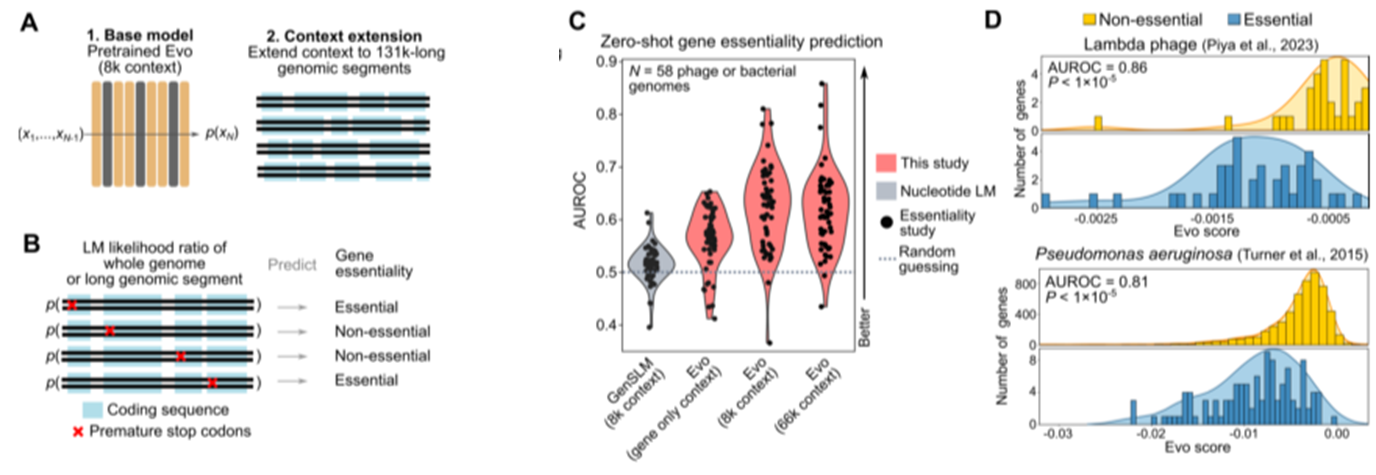
Generating DNA sequences at genome scale
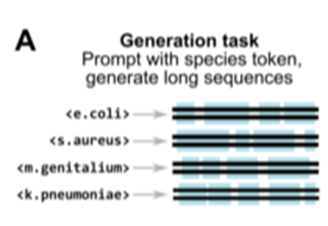
Synthetic sequence generation
- prompted with species-level tokens during the second pretraining
- bacterial species promts -> generate seuqnces of ~650kb in length
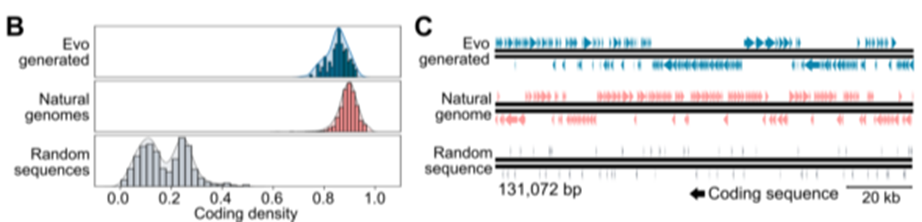
Evaluation
- depicts the organization of coding sequences
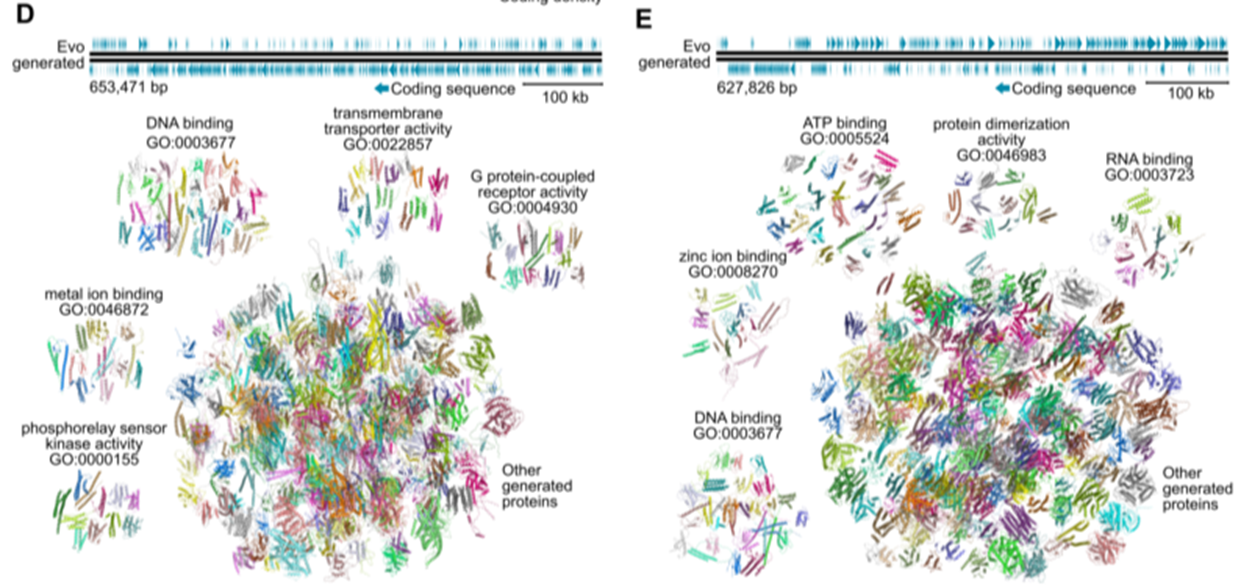
ESMFold structure predictions
Usage
Paper and GitHub Repository You can use Evo model in Together AI
Discussion
Safety and ethics discussion
- whole-genome foundation models have thepotential for misuse
- threat to biosafety and biosecurity?
- can also catalyze the development of harmful synthetic microorganisms
- whole-genome foundation models could contribute to social and health inequity
- companies may accelerate research that prioritizes returns-on-investment over the global disease burden or health equity
- may enable an organization to bypass current intellectual property
- whole-genome foundation models could contribute to disruptions to the natural environment
- intellectual property law should evolve as generative models increasingly automate the biological discovery and design process
The path forward
- establishment of clear, comprehensive guidelines that delineate ethical practices
- community partnerships and international collaborations – address disparities in access and capabilities
- create a dynamic feedback loop that engages all share-holders in a continuous dialogue
-> lays the groundwork for a future where genetic engineering advances in harmony with ethical principles and societal values

Leave a comment