[scRNA-seq] ssGSEA with escape R package in Seurat v3 and later versions - troubleshooting
The most recent version of Seurat is v5.1.0
In Seruat v5, the seurat object was changed to a layer structure, so to access the assay data in Seurat object, you need to use the code like:
<obj>[["RNA"]]$counts
or
LayerData(<obj>, assay = "RNA", layer = "counts")
or
GetAssayData(object = pbmc, assay = "RNA", slot = "counts")
Instead, the following code is now not available, which was the code for the versions prior to v3.
<obj>$assays$RNA@counts
However, when using the enrichIT function in the escape package, the code still uses
So I redefined the functions by hand and wrote below.
One thing that I’ll update later is performing parallel computing using BiocParallel.
# ssGSEA
p_load(escape, dittoSeq, ssGSEA)
options("Seurat.object.assay.version" = "v3")
DefaultAssay(mEpC) <- "RNA"
GS.hallmark <- getGeneSets(species = "Mus musculus", library = "H")
p_load(BiocParallel, snow)
performPCA <- function(enriched, gene.sets = NULL, groups) {
groups <- data.frame(enriched[,colnames(enriched) %in% c(groups)])
input <- select_if(enriched, is.numeric)
if (!is.null(gene.sets)) {
input <- input[,colnames(input) %in% gene.sets]
}
PCA <- prcomp(input, scale. = TRUE)
merged <- cbind(PCA$x, groups)
return(merged)
}
split_data.matrix <- function(matrix, chunk.size=1000) {
ncols <- dim(matrix)[2]
nchunks <- (ncols-1) %/% chunk.size + 1
split.data <- list()
min <- 1
for (i in seq_len(nchunks)) {
if (i == nchunks-1) { #make last two chunks of equal size
left <- ncols-(i-1)*chunk.size
max <- min+round(left/2)-1
} else {
max <- min(i*chunk.size, ncols)
}
split.data[[i]] <- matrix[,min:max]
min <- max+1 #for next chunk
}
return(split.data)
}
getGeneSets <- function(species = "Homo sapiens",
library = NULL,
subcategory = NULL,
gene.sets = NULL) {
spec <- msigdbr_species()
spec_check <- unlist(spec[spec$species_name %in% species,][,1])
if (length(spec_check) == 0) {
message(paste0("Please select a compatible species: ",
paste(spec, collapse = ", ")))
}
if(!is.null(library)) {
if (length(library) == 1) {
if (is.null(subcategory)) {
m_df = msigdbr(species = spec_check, category = library)
} else {
m_df = msigdbr(species = spec_check, category = library, subcategory = subcategory)
}
}
m_df <- NULL
for (x in seq_along(library)) {
if (is.null(subcategory)) {
tmp2 = msigdbr(species = spec_check, category = library[x])
} else {
tmp2 = msigdbr(species = spec_check, category = library, subcategory = subcategory)
}
m_df <- rbind(m_df, tmp2)
}
if(!is.null(gene.sets)) {
m_df <- m_df[m_df$gs_name %in% gene.sets,]
}
}
gs <- unique(m_df$gs_name)
ls <- list()
for (i in seq_along(gs)) {
tmp <- m_df[m_df$gs_name == gs[i],]
tmp <- tmp$gene_symbol
tmp <- unique(tmp)
tmp <- GeneSet(tmp, setName=paste(gs[i]))
ls[[i]] <- tmp
}
gsc <- GeneSetCollection(ls)
return(gsc)
}
normalize <- function(x)
{
(x- min(x)) /(max(x)-min(x))
}
GS.check <- function(gene.sets) {
if(is.null(gene.sets)) {
stop("Please provide the gene.sets you would like to use for
the enrichment analysis")
}
egc <- gene.sets
if(inherits(egc, what = "GeneSetCollection")){
egc <- GSEABase::geneIds(egc) # will return a simple list,
#which will work if a matrix is supplied to GSVA
}
return(egc)
}
cntEval2 <- function(obj) {
if (inherits(x = obj, what = "Seurat")) {
cnts <- GetAssayData(object = obj, assay = "RNA", slot = "counts")
} else if (inherits(x = obj, what = "SingleCellExperiment")) {
cnts <- counts(obj)
} else {
cnts <- obj
}
if (!inherits(cnts, what = "dgCMatrix")) {
cnts <- Matrix(as.matrix(cnts),sparse = TRUE)
}
cnts <- cnts[tabulate(summary(cnts)$i) != 0, , drop = FALSE]
return(cnts)
}
enrichIt2 <- function (obj, gene.sets = NULL, method = "ssGSEA", groups = 1000,
cores = 2, min.size = 5, ssGSEA.norm = FALSE, ...)
{
library(escape)
egc <- GS.check(gene.sets)
cnts <- cntEval2(obj)
if (!is.null(min.size)) {
GS.size <- lapply(egc, function(x) length(which(rownames(cnts) %in%
x)))
remove <- unname(which(GS.size < min.size))
if (length(remove) != 0) {
egc <- egc[-remove]
}
}
scores <- list()
wind <- seq(1, ncol(cnts), by = groups)
print(paste("Using sets of", groups, "cells. Running", length(wind),
"times."))
if (method == "ssGSEA") {
split.data <- split_data.matrix(matrix = cnts, chunk.size = groups)
for (i in seq_along(wind)) {
last <- min(ncol(cnts), i + groups - 1)
a <- suppressWarnings(gsva(split.data[[i]], egc,
method = "ssgsea", ssgsea.norm = FALSE, kcdf = "Poisson"#,
#parallel.sz = cores, BPPARAM = SnowParam()
),
...)
scores[[i]] <- a
}
}
else if (method == "UCell") {
scores[[1]] <- t(suppressWarnings(ScoreSignatures_UCell(cnts,
features = egc, chunk.size = groups, ncores = cores,
...)))
}
scores <- do.call(cbind, scores)
output <- t(as.matrix(scores))
if (method == "ssGSEA" & ssGSEA.norm) {
output <- apply(output, 2, normalize)
}
output <- data.frame(output)
return(output)
}
add.meta.data <- function(sc, meta, header) {
if (inherits(x=sc, what ="Seurat")) {
col.name <- names(meta) %||% colnames(meta)
sc[[col.name]] <- meta
} else {
rownames <- rownames(colData(sc))
colData(sc) <- cbind(colData(sc),
meta[rownames,])[, union(colnames(colData(sc)), colnames(meta))]
rownames(colData(sc)) <- rownames
}
return(sc)
}
Now we can perform ssGSEA with these functions defined above.
ss.mEpC <- enrichIt2(obj = mEpC, gene.sets = GS.hallmark, groups= 1000, min.size = 5)
mEpC <- AddMetaData(mEpC, ss.mEpC)
p_load(magrittr, ArchR, grid, viridis)
devtools::install_github("GreenleafLab/ArchR"); library(ArchR)
my_palette <- colorRampPalette(c('blue', '#F5F5F5', 'red'))(100)
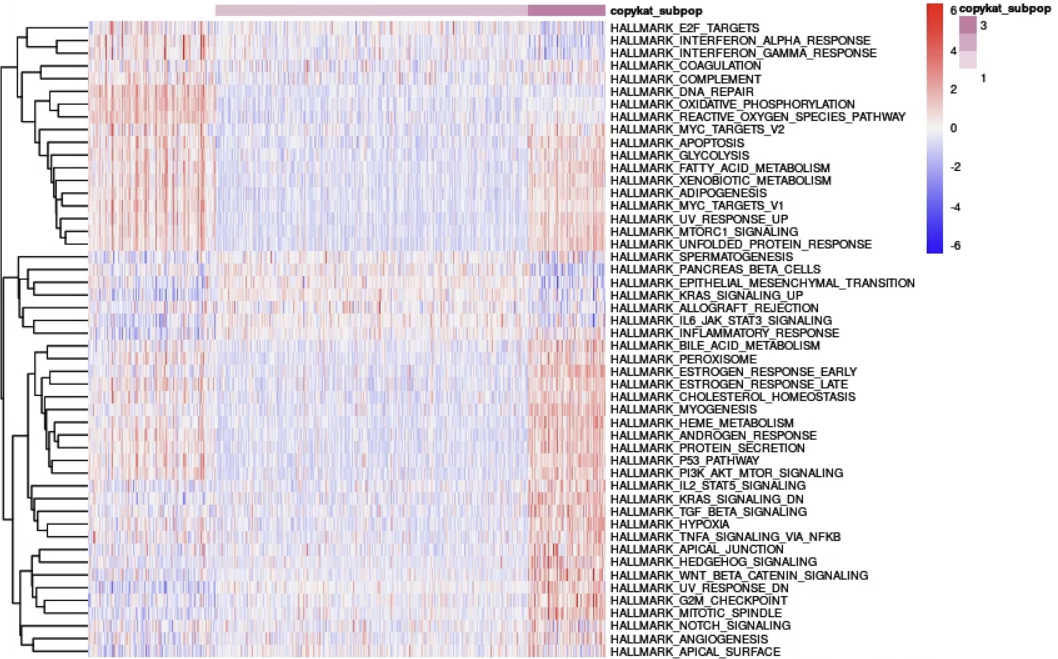
dittoHeatmap(mEpC,
genes = NULL, metas = names(ss.mEpC),
annot.by = "copykat_subpop",
fontsize = 7,
cluster_cols = FALSE,
heatmap.colors = my_palette)
And you can get a result like this.

Leave a comment