CellRank 2: A Unified Framework for Single-Cell Fate Mapping
CellRank 2: Revolutionizing Single-Cell Trajectory Analysis
In a groundbreaking study published in Nature Methods, Weiler et al. introduce CellRank 2, a powerful and versatile computational framework for analyzing cellular trajectories and fate decisions in single-cell data. This innovative tool represents a significant advancement in the field of single-cell biology, offering a unified approach to trajectory inference across diverse data modalities.
| (Paper | Code) |
Key Features of CellRank 2:
- Modular architecture with kernels and estimators for flexible analysis
- Ability to integrate multiple data types including RNA velocity, pseudotime, and metabolic labeling
- Enhanced performance, capable of analyzing datasets with over a million cells
- Advanced methods for identifying terminal states, calculating fate probabilities, and pinpointing driver genes
The paper demonstrates CellRank 2’s capabilities through applications in various biological systems, including human hematopoiesis, embryoid body development, and intestinal organoid differentiation. By providing a standardized, scalable approach to trajectory analysis, CellRank 2 promises to accelerate discoveries in developmental biology, regenerative medicine, and cancer research.
In this blog post, we’ll dive deep into the technical innovations behind CellRank 2, explore its applications, and discuss its potential impact on the future of single-cell biology research.
Introduction
In the rapidly evolving field of single-cell biology, understanding cellular trajectories and fate decisions is crucial for deciphering complex biological processes. However, the diversity of single-cell data types and the computational challenges associated with analyzing large-scale datasets have long been significant hurdles. Enter CellRank 2, a groundbreaking framework that promises to revolutionize how we approach single-cell trajectory inference.
CellRank 2, developed by Weiler et al. and published in Nature Methods, builds upon its predecessor to offer a unified, flexible, and scalable solution for analyzing cellular dynamics across various data modalities. This new iteration addresses several key challenges in the field:
-
Data Diversity: With the proliferation of single-cell technologies, researchers now have access to a wide array of data types, including RNA velocity, pseudotime orderings, and metabolic labeling. CellRank 2 provides a cohesive framework to integrate and analyze these diverse data sources.
-
Scalability: As dataset sizes continue to grow, reaching millions of cells, traditional methods often falter. CellRank 2 introduces optimizations that allow it to handle these massive datasets efficiently.
-
Flexibility: Biological systems are diverse, and no single analytical approach fits all scenarios. CellRank 2’s modular design allows researchers to tailor their analyses to the specific needs of their study.
-
Interpretability: Beyond just identifying trajectories, understanding the molecular drivers of fate decisions is crucial. CellRank 2 provides tools to identify putative driver genes and visualize gene expression trends along trajectories.
By addressing these challenges, CellRank 2 positions itself as a comprehensive solution for researchers seeking to unravel the complexities of cellular differentiation, development, and reprogramming. In this blog post, we’ll dive deep into the technical innovations that power CellRank 2, explore its applications across various biological systems, and discuss its potential impact on the field of single-cell biology.
Key Innovations of CellRank 2
CellRank 2 introduces several groundbreaking features that set it apart from its predecessor and other trajectory inference methods. Let’s dive into these key innovations:
1. Modular Architecture
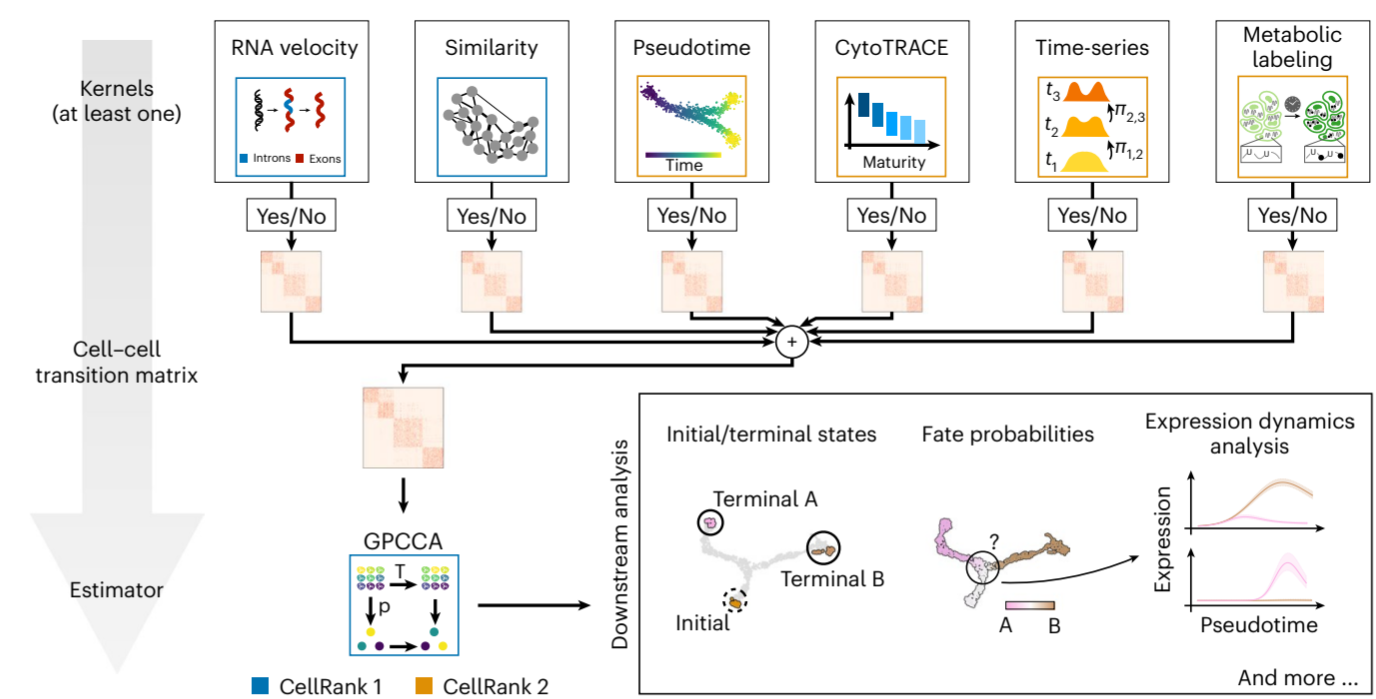 [Figure placeholder - Modular architecture of CellRank 2, adapted from Fig. 1 in the manuscript]
[Figure placeholder - Modular architecture of CellRank 2, adapted from Fig. 1 in the manuscript]
The cornerstone of CellRank 2’s flexibility is its modular architecture, which separates the analysis pipeline into two main components:
- Kernels: These modules estimate cell-cell transition probabilities from various data types.
- Estimators: These analyze the transition probabilities to derive biological insights.
This separation allows researchers to mix and match different data sources and analysis techniques, tailoring the approach to their specific biological question and data types.
2. New Kernels for Multiple Data Modalities
CellRank 2 expands its applicability by introducing new kernels to handle diverse data types:
- PseudotimeKernel: Leverages pseudotime information to infer cellular dynamics.
- CytoTRACEKernel: Utilizes developmental potential estimates to reconstruct trajectories.
- RealTimeKernel: Incorporates experimental time points, allowing for the analysis of time-series data.
- Metabolic Labeling Integration: A new method to infer kinetic rates and cellular dynamics from metabolic labeling experiments.
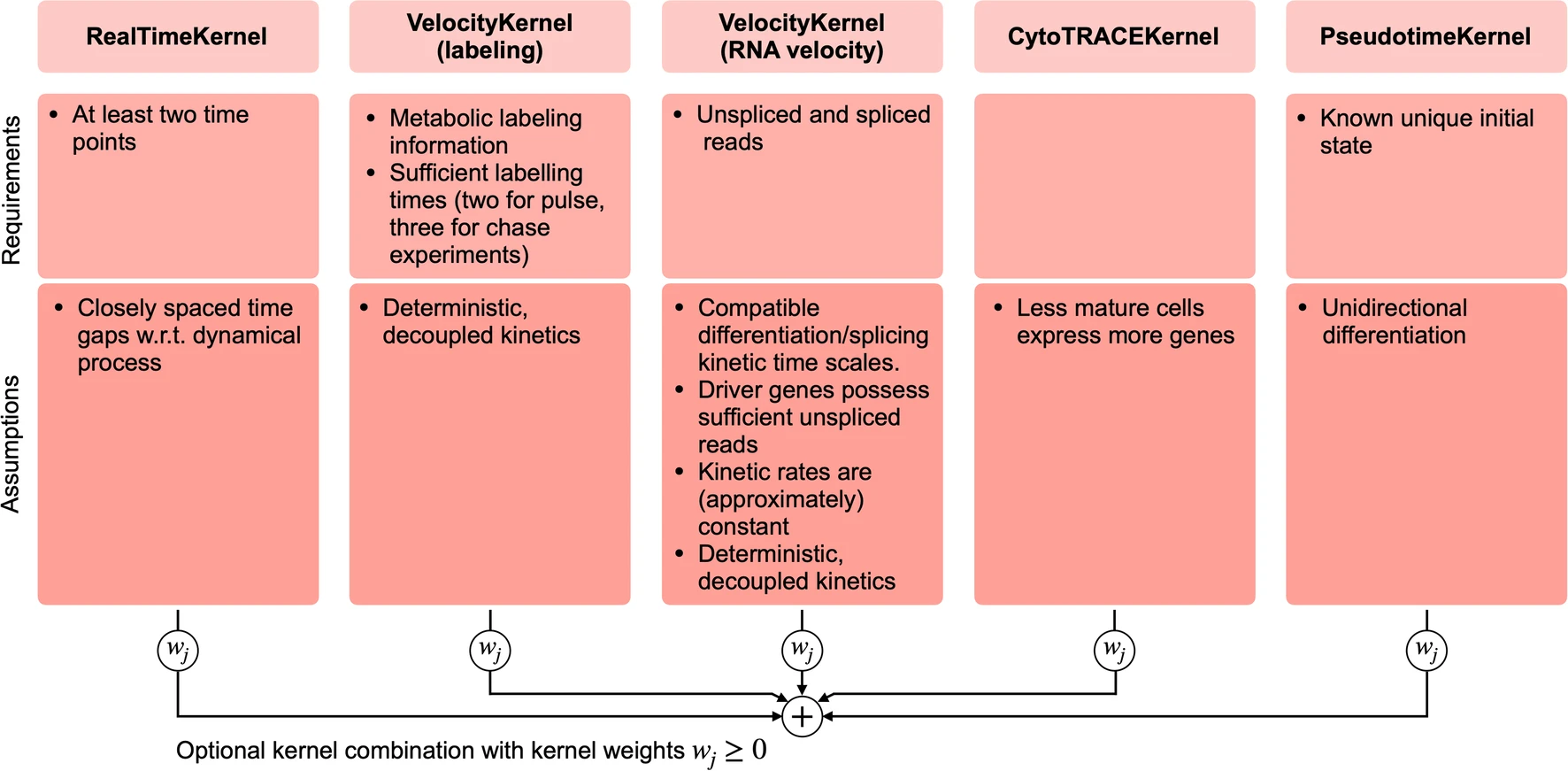 [Figure placeholder - Comparison of different kernels, similar to Extended Data Fig. 1 in the manuscript]
[Figure placeholder - Comparison of different kernels, similar to Extended Data Fig. 1 in the manuscript]
These new kernels significantly broaden the scope of data that can be analyzed within a unified framework, enabling more comprehensive insights into cellular fate decisions.
3. Performance Improvements
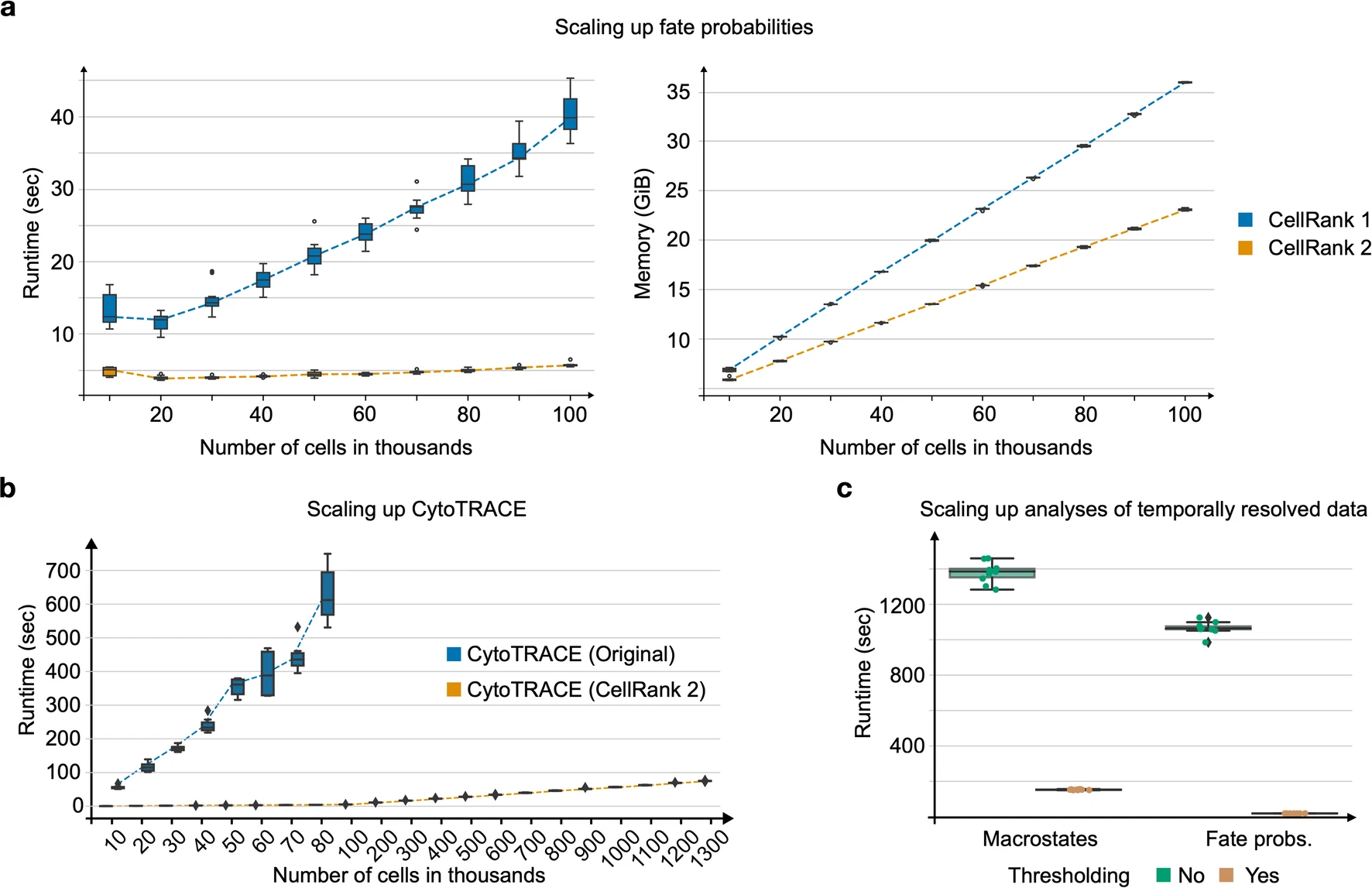 [Figure placeholder - Performance comparison, adapted from Extended Data Fig. 2 in the manuscript]
[Figure placeholder - Performance comparison, adapted from Extended Data Fig. 2 in the manuscript]
CellRank 2 introduces substantial performance enhancements:
- Scalability: Can handle datasets of over a million cells, a significant improvement over its predecessor.
- Computational Optimizations: Includes a 30-fold speed-up in fate probability calculations and memory usage optimizations.
- Adaptive Thresholding: A new scheme for sparse transition matrices, enabling faster downstream computations.
These improvements make CellRank 2 suitable for the analysis of increasingly large and complex single-cell datasets.
4. Enhanced Visualization and Interpretation Tools
CellRank 2 provides several new tools to aid in the interpretation of results:
- Random Walk Visualization: Allows for qualitative understanding of cellular dynamics.
- Real-time Informed Pseudotime: A new approach that combines experimental time points with continuous expression changes.
- Improved Driver Gene Identification: Enhanced methods for identifying genes that may be driving fate decisions.
These tools help researchers not only to identify trajectories but also to understand the underlying molecular mechanisms driving cellular fate decisions.
By combining these innovations, CellRank 2 provides a powerful, flexible, and user-friendly framework for analyzing cellular trajectories across a wide range of biological systems and experimental designs.
Technical Deep Dive
To fully appreciate the power and flexibility of CellRank 2, let’s delve into its technical underpinnings. This section will explore the mathematical framework, kernel-specific algorithms, and estimator functionalities that form the core of CellRank 2.
1. Mathematical Framework
At its heart, CellRank 2 uses a Markov chain model to describe cellular dynamics probabilistically. This approach allows for a robust, noise-tolerant representation of cell state transitions.
Markov Chain Modeling
In CellRank 2, each cell represents a state in a Markov chain. The transition probabilities between states (cells) are encoded in a transition matrix T ∈ ℝ^(n_c × n_c), where n_c is the number of cells. Each row T_j,: represents the transition probabilities of cell j to its potential descendants.
 [Figure placeholder - Visualization of Markov chain modeling, could be adapted from Fig. 1 in the manuscript]
[Figure placeholder - Visualization of Markov chain modeling, could be adapted from Fig. 1 in the manuscript]
This probabilistic framework allows CellRank 2 to capture the gradual and noisy nature of fate decisions in biological systems.
2. Kernel-Specific Algorithms
CellRank 2 introduces several new kernels, each with its own algorithm for computing transition probabilities. Let’s examine the key ones:
PseudotimeKernel
This kernel biases a cell-cell similarity graph based on pseudotime values. For cells j and k with pseudotime difference Δt_jk, the edge weight C_jk is updated as:
C_jk = C̃_jk * f(Δt_jk)
Where C̃_jk is the original similarity-based edge weight, and f is a thresholding function. CellRank 2 offers both soft and hard thresholding options.
RealTimeKernel
This kernel combines inter-time point transitions (computed using optimal transport) with intra-time point transitions based on transcriptomic similarity. The global transition matrix T incorporates both:
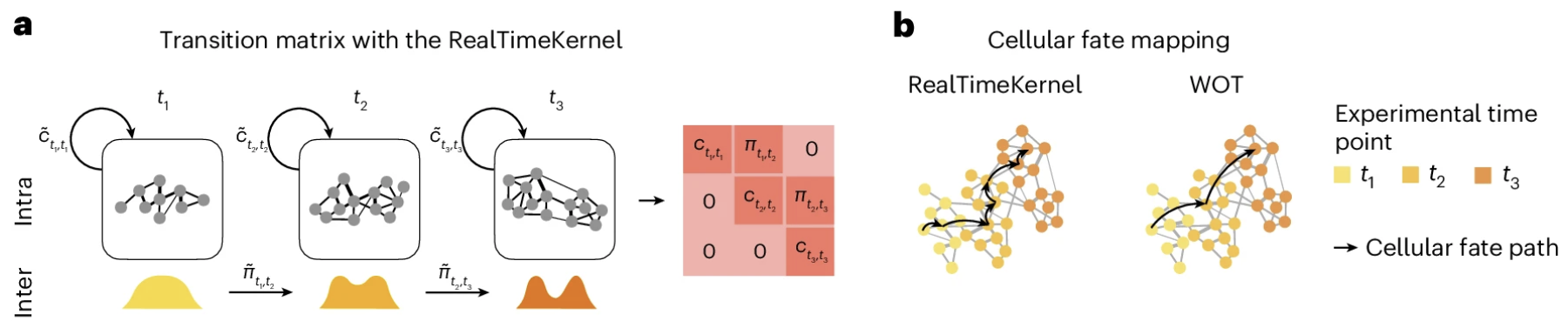 [Figure placeholder - Illustration of RealTimeKernel, adapted from Fig. 4a-b in the manuscript]
[Figure placeholder - Illustration of RealTimeKernel, adapted from Fig. 4a-b in the manuscript]
Metabolic Labeling Integration
For metabolic labeling data, CellRank 2 estimates cell-specific transcription (α) and degradation (γ) rates by modeling mRNA dynamics:
ṙ = α - γr
Where r is the mRNA level. Parameters are estimated by minimizing a quadratic loss function across different labeling durations.
3. Estimator Functionalities
Once transition probabilities are computed, CellRank 2’s estimators perform various analyses:
Initial and Terminal State Identification
CellRank 2 uses the generalized Perron cluster cluster analysis (GPCCA) method to identify macrostates from the transition matrix. These are then classified as initial, intermediate, or terminal states.
Fate Probability Calculation
Fate probabilities toward terminal states are computed using absorption probabilities in the Markov chain. CellRank 2 introduces a new, faster method for this calculation:
Gene Expression Trend Analysis
CellRank 2 enables the visualization of gene expression trends along trajectories and the identification of genes correlated with specific fates. This is achieved by fitting generalized additive models to describe gene expression changes over pseudotime.
 [Figure placeholder - Example of gene expression trend analysis, could be adapted from Fig. 3 in the manuscript]
[Figure placeholder - Example of gene expression trend analysis, could be adapted from Fig. 3 in the manuscript]
By combining these sophisticated mathematical techniques with efficient computational implementations, CellRank 2 provides a powerful and flexible framework for analyzing cellular trajectories in diverse biological contexts.
Applications and Case Studies
CellRank 2’s versatility and power are best demonstrated through its successful application to diverse biological systems. In this section, we’ll explore five key case studies that showcase the framework’s capabilities across different data modalities and biological contexts.
1. Human Hematopoiesis
In this study, CellRank 2 was applied to analyze human hematopoiesis using the PseudotimeKernel.
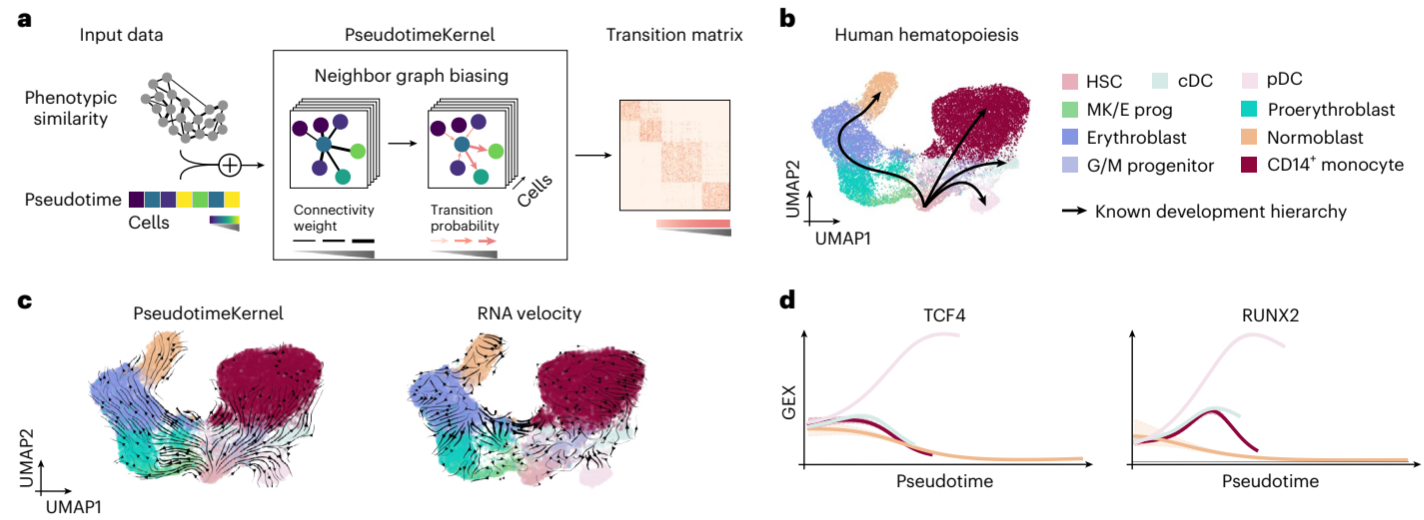 [Figure placeholder - UMAP of hematopoiesis data, adapted from Fig. 2 in the manuscript]
[Figure placeholder - UMAP of hematopoiesis data, adapted from Fig. 2 in the manuscript]
Key findings:
- Successfully recovered all four terminal states and the initial state.
- Identified known lineage drivers for the plasmacytoid dendritic cell (pDC) lineage, including RUNX2 and TCF4.
- Outperformed RNA velocity-based analysis, which failed to recover the classical dendritic cell (cDC) lineage.
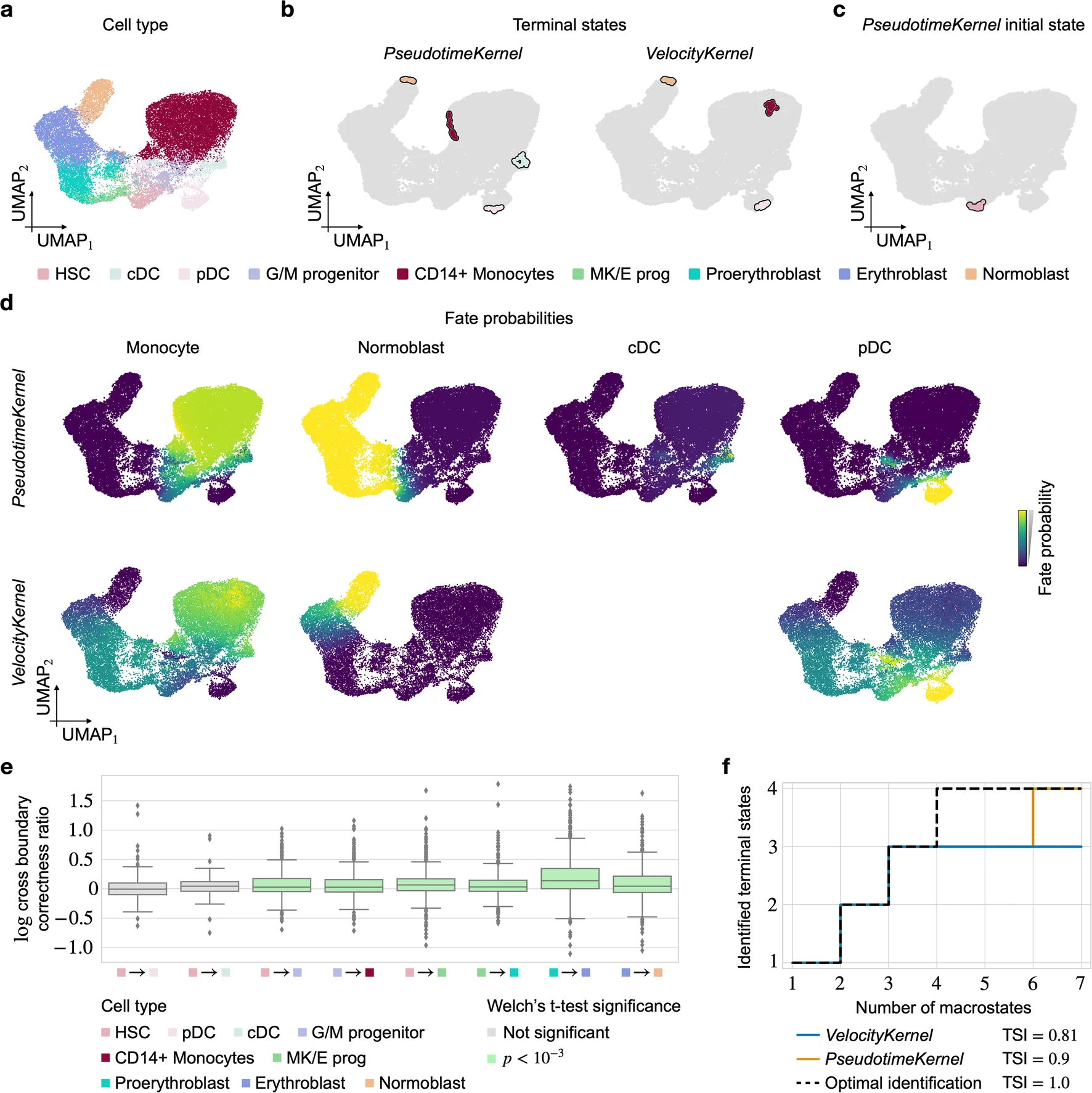 [Figure placeholder - Comparison of PseudotimeKernel and VelocityKernel results, adapted from Extended Data Fig. 3 in the manuscript]
[Figure placeholder - Comparison of PseudotimeKernel and VelocityKernel results, adapted from Extended Data Fig. 3 in the manuscript]
This case study demonstrates CellRank 2’s ability to overcome limitations of RNA velocity in steady-state systems.
2. Embryoid Body Development
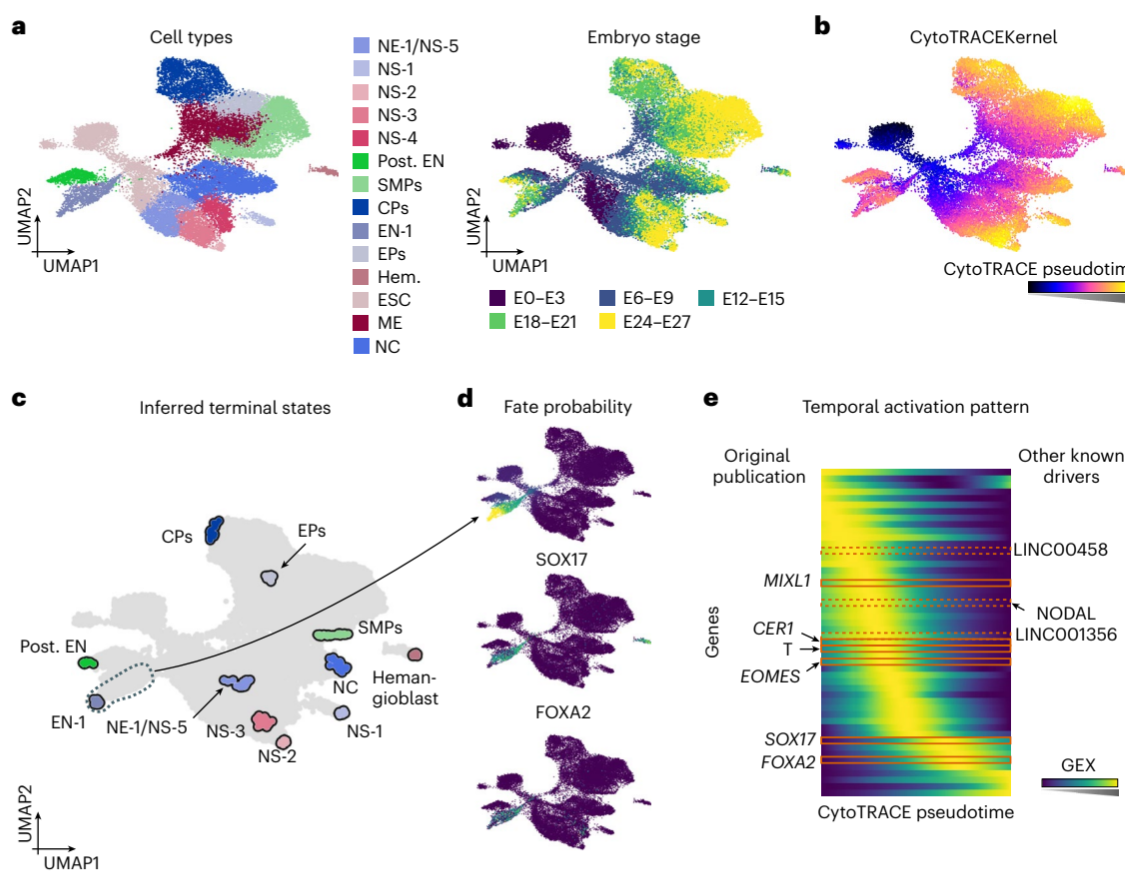
For this application, CellRank 2 utilized the CytoTRACEKernel to study endoderm development in embryoid bodies.
 [Figure placeholder - UMAP of embryoid body data, adapted from Fig. 3 in the manuscript]
[Figure placeholder - UMAP of embryoid body data, adapted from Fig. 3 in the manuscript]
Key outcomes:
- Identified 10 out of 11 terminal cell populations and the correct initial state.
- Recovered known driver genes of endoderm development, including MIXL1, FOXA2, and SOX17.
- Revealed the temporal activation pattern of key genes during endoderm development.
This study showcases CellRank 2’s ability to infer cellular dynamics without specifying an initial state for pseudotime computation.
3. Mouse Embryonic Fibroblast (MEF) Reprogramming
CellRank 2’s RealTimeKernel was employed to analyze MEF reprogramming data, incorporating both experimental time points and intra-time point dynamics.
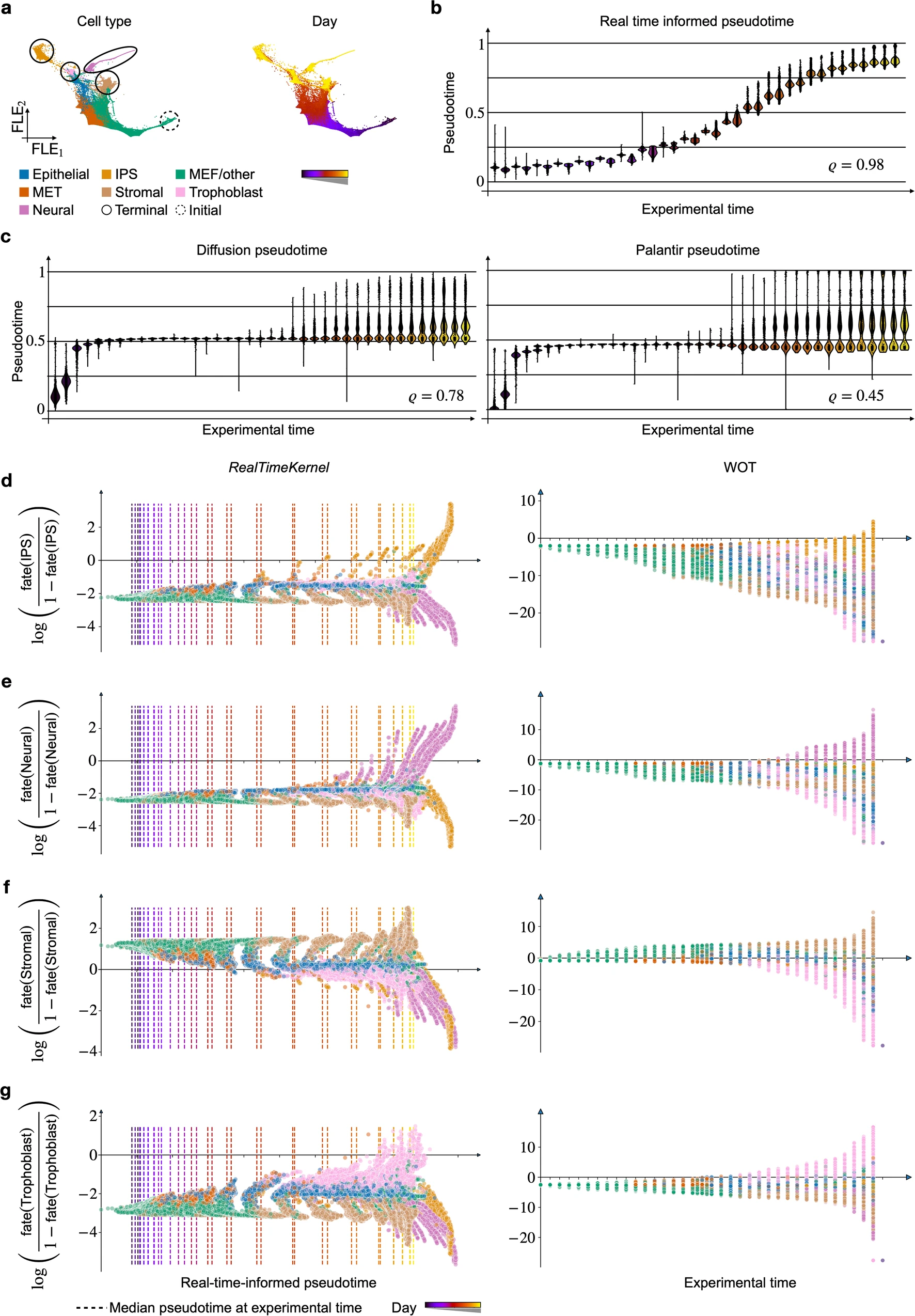 [Figure placeholder - FLE of MEF reprogramming data, adapted from Extended Data Fig. 7 in the manuscript]
[Figure placeholder - FLE of MEF reprogramming data, adapted from Extended Data Fig. 7 in the manuscript]
Highlights:
- Successfully modeled fate decisions on a continuous domain.
- Provided a more granular mapping of cell fate compared to traditional optimal transport methods.
- Enabled studying gradual fate establishment along a continuous axis.
This case demonstrates the power of combining inter- and intra-time point information for a more complete picture of cellular dynamics.
4. Pharyngeal Endoderm Development
The RealTimeKernel was applied to study pharyngeal endoderm development from embryonic days 9.5 to 12.5.
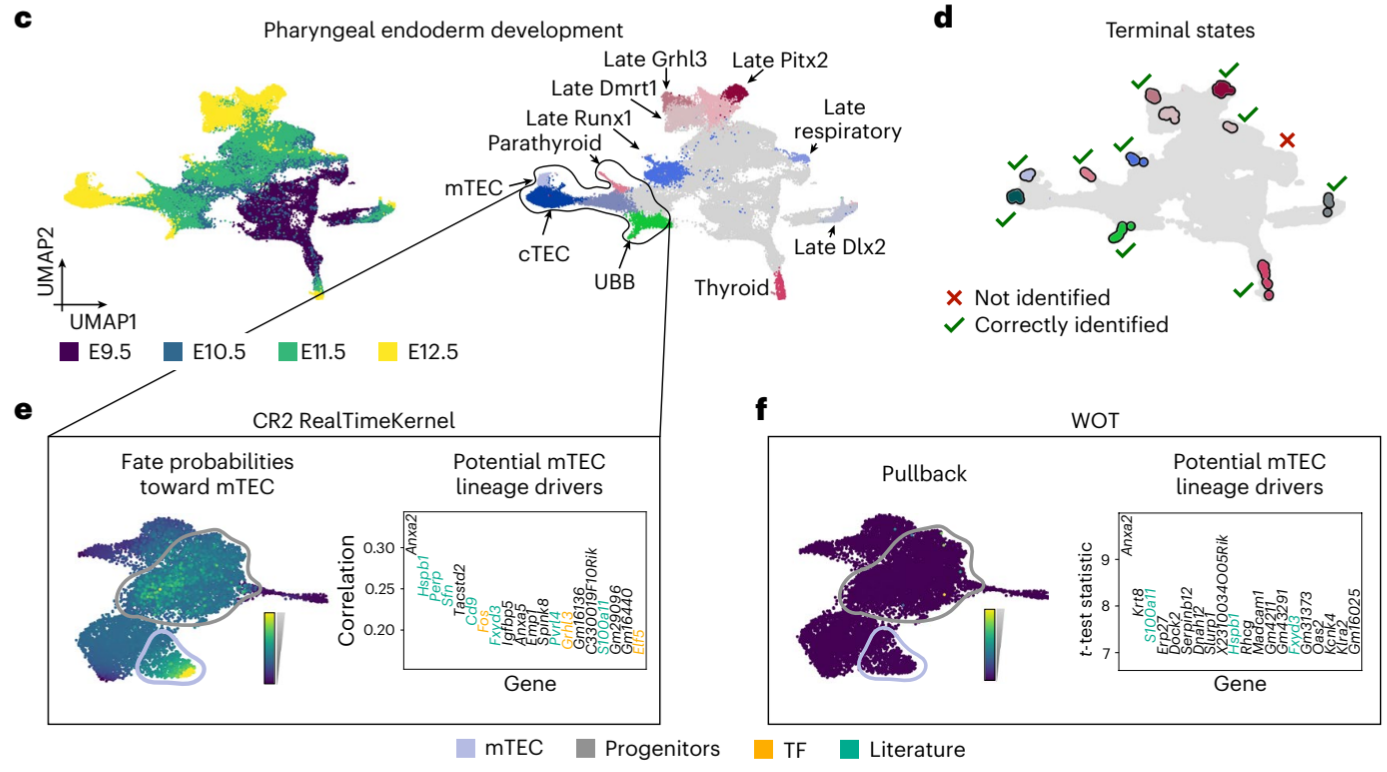 [Figure placeholder - UMAP of pharyngeal endoderm data, adapted from Fig. 4c-f in the manuscript]
[Figure placeholder - UMAP of pharyngeal endoderm data, adapted from Fig. 4c-f in the manuscript]
Key results:
- Recovered 10 out of 11 terminal states manually assigned in the original publication.
- Identified known lineage drivers for parathyroid (Gcm2), thyroid (Hhex), and thymus (Foxn1) development.
- Discovered a putative progenitor population for medullary thymic epithelial cells (mTECs).
This study highlights CellRank 2’s ability to uncover novel biological insights in complex developmental processes.
5. Intestinal Organoid Differentiation
For this application, CellRank 2 integrated metabolic labeling data to study intestinal organoid differentiation.
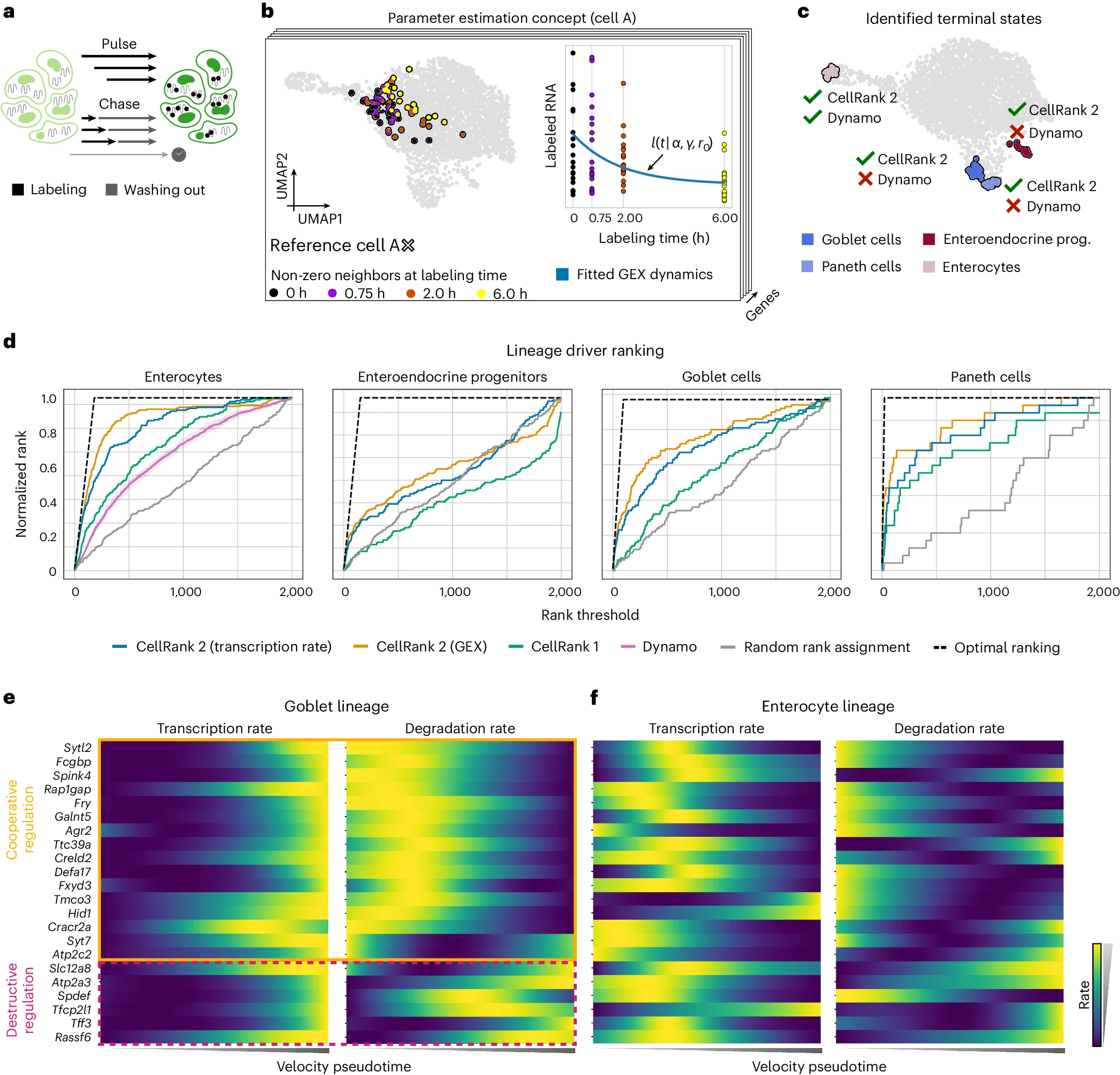 [Figure placeholder - UMAP of intestinal organoid data, adapted from Fig. 5 in the manuscript]
[Figure placeholder - UMAP of intestinal organoid data, adapted from Fig. 5 in the manuscript]
Key findings:
- Successfully recovered all four terminal states with high cell-type purity.
- Outperformed classical RNA velocity in ranking known lineage drivers.
- Revealed different regulatory strategies for lineage-correlated genes.
This case study demonstrates CellRank 2’s unique ability to leverage metabolic labeling data for inferring cellular dynamics and regulatory strategies.
These diverse applications underscore CellRank 2’s versatility and power in analyzing cellular trajectories across various biological systems and data modalities. By providing a unified framework for these analyses, CellRank 2 enables researchers to gain deeper insights into complex biological processes.
Comparative Analysis
To fully appreciate the advancements brought by CellRank 2, it’s crucial to compare its performance against other established methods in the field. This section provides a comprehensive comparison of CellRank 2 with alternative approaches across various metrics and use cases.
1. Performance and Scalability
CellRank 2 demonstrates significant improvements in computational efficiency compared to its predecessor and other methods.
Key points:
- CellRank 2 shows a 30-fold speed-up in fate probability calculations compared to the original CellRank.
- It can handle datasets of over 1 million cells, a substantial improvement over many existing methods.
- Memory usage is significantly reduced, allowing for analysis of larger datasets on standard hardware.
2. Terminal State Identification
The accuracy of terminal state identification is a critical metric for trajectory inference methods. CellRank 2 consistently outperforms other methods in this aspect.
[Figure placeholder - Terminal State Identification (TSI) score comparison, adapted from Extended Data Fig. 3f, 6d, 8b, and 10d in the manuscript]
Highlights:
- In the hematopoiesis dataset, CellRank 2’s PseudotimeKernel achieved a TSI score of 0.9, compared to 0.81 for the RNA velocity-based approach.
- For the pharyngeal endoderm development data, the RealTimeKernel scored a TSI of 0.92, significantly higher than the VelocityKernel’s 0.46.
- In the intestinal organoid study, CellRank 2 outperformed CellRank 1 with TSI scores of 0.81 and 0.71, respectively.
3. Driver Gene Identification
CellRank 2’s ability to identify potential driver genes of cellular fate decisions is compared with other methods, including dynamo and classical differential expression analysis.
Key findings:
- In the intestinal organoid study, CellRank 2 consistently outperformed competing approaches in ranking known lineage drivers for all four terminal states.
- CellRank 2’s correlation-based analysis identified more relevant drivers compared to classical differential expression testing in the pharyngeal endoderm development study.
4. Handling of Time-Course Data
CellRank 2’s RealTimeKernel is compared with Waddington-OT (WOT) for analyzing time-course data.
Notable differences:
- The RealTimeKernel identified a putative mTEC ancestor cluster in the pharyngeal endoderm data, which WOT failed to detect.
- CellRank 2 recovered substantially more relevant drivers compared to WOT, likely due to its incorporation of both inter- and intra-time point dynamics.
5. Metabolic Labeling Data Analysis
CellRank 2’s approach to analyzing metabolic labeling data is compared with dynamo, a recently developed method for this data type.
Key differences:
- CellRank 2 successfully recovered all four terminal states in the intestinal organoid data, while dynamo only identified the enterocyte population.
- CellRank 2 estimates cell-specific transcription and degradation rates, providing a more granular view of cellular dynamics compared to dynamo’s assumptions of constant rates across cells.
6. Flexibility and Adaptability
While harder to quantify, CellRank 2’s modular design provides significant advantages in terms of flexibility:
- It can easily incorporate new data types through the addition of new kernels.
- The separation of kernels and estimators allows for easy combination of different data modalities.
- This adaptability makes CellRank 2 well-suited for the rapidly evolving landscape of single-cell technologies.
In summary, CellRank 2 demonstrates superior performance across multiple metrics and use cases. Its combination of computational efficiency, accuracy in trajectory inference, and flexibility in handling diverse data types positions it as a leading tool in the field of single-cell trajectory analysis.
Future Directions and Potential Impact
The introduction of CellRank 2 marks a significant advancement in single-cell trajectory analysis. However, science is always evolving, and there are exciting possibilities for further development and application of this framework. In this section, we’ll explore potential future directions for CellRank 2 and its broader impact on the field of single-cell biology.
1. Integration with Emerging Single-Cell Technologies
As single-cell technologies continue to advance, CellRank 2 is well-positioned to adapt and incorporate new data types:
- Spatial Transcriptomics: The integration of spatial information could provide insights into how cellular microenvironments influence fate decisions.
- Multi-omics Integration: Combining transcriptomics with epigenomics or proteomics data could offer a more comprehensive view of cellular states and transitions.
- Single-Cell Lineage Tracing: Incorporating genetic lineage tracing data could further refine trajectory inference.
2. Enhanced Causal Inference
While CellRank 2 excels at identifying putative driver genes, future versions could incorporate more sophisticated causal inference methods:
- Integration with Perturbation Data: Combining CellRank 2’s trajectory analysis with single-cell perturbation experiments could help establish causal relationships between gene expression and fate decisions.
- Causal Network Inference: Developing methods to infer causal gene regulatory networks along trajectories could provide deeper insights into the mechanisms of cellular differentiation.
3. Dynamic Kernel Combinations
Currently, CellRank 2 allows for global weighting of different kernels. Future developments could include:
- Local Kernel Combinations: Allowing kernel weights to vary based on the position of cells within the phenotypic manifold, enabling context-dependent integration of multiple data sources.
- Automated Kernel Selection: Developing methods to automatically determine the most appropriate kernel(s) for a given dataset based on its characteristics.
4. Expansion to New Biological Systems
While CellRank 2 has been successfully applied to various systems, there’s potential for expansion:
- Cancer Progression: Applying CellRank 2 to study tumor evolution and metastasis.
- Cellular Reprogramming: Further refinement of the framework for studying induced pluripotency and transdifferentiation.
- Aging Studies: Investigating cellular trajectory changes associated with aging.
5. Improved Visualization and Interpretation Tools
Building on CellRank 2’s existing visualization capabilities:
- Interactive Visualization: Developing interactive tools for exploring trajectories and gene expression trends.
- 3D Trajectory Visualization: Creating methods for visualizing complex trajectories in three-dimensional space.
6. Integration with Machine Learning and AI
As AI continues to advance, there are opportunities to enhance CellRank 2:
- Deep Learning for Feature Extraction: Using deep learning models to extract relevant features for trajectory inference.
- Reinforcement Learning for Parameter Optimization: Developing RL algorithms to optimize kernel parameters and combinations.
Potential Impact
The continued development and application of CellRank 2 could have far-reaching impacts:
-
Accelerating Developmental Biology Research: By providing a unified framework for analyzing diverse single-cell data types, CellRank 2 could speed up discoveries in developmental biology.
-
Advancing Regenerative Medicine: Improved understanding of cellular differentiation trajectories could inform strategies for tissue engineering and regenerative therapies.
-
Enhancing Cancer Research: Better characterization of tumor evolution could lead to new therapeutic strategies targeting specific cellular states along cancer progression trajectories.
-
Facilitating Drug Discovery: Understanding cellular state transitions could help identify new drug targets and predict drug responses at a single-cell level.
-
Standardizing Trajectory Analysis: As CellRank 2 becomes more widely adopted, it could help standardize approaches to trajectory inference across the field, facilitating comparisons between studies.
In conclusion, CellRank 2 not only represents a significant advance in our current capabilities for single-cell trajectory analysis but also lays the groundwork for exciting future developments. Its flexible and extensible framework positions it to adapt to new technologies and drive discoveries across various areas of biological research.
Conclusion
The introduction of CellRank 2 marks a significant milestone in the field of single-cell biology, particularly in the realm of cellular trajectory analysis. As we’ve explored throughout this blog post, CellRank 2 brings several key advancements to the table:
-
Unified Framework: By providing a modular architecture with kernels and estimators, CellRank 2 offers a unified approach to analyzing diverse single-cell data types. This flexibility allows researchers to tackle a wide range of biological questions using a single, coherent framework.
-
Enhanced Performance: With its improved computational efficiency and ability to handle datasets of over a million cells, CellRank 2 is well-equipped to meet the demands of modern single-cell studies.
-
Diverse Applications: From human hematopoiesis to intestinal organoid differentiation, CellRank 2 has demonstrated its versatility across various biological systems and experimental designs.
-
Integration of Multiple Data Modalities: The framework’s ability to incorporate RNA velocity, pseudotime, developmental potential, experimental time points, and metabolic labeling data provides a more comprehensive view of cellular dynamics.
-
Robust Analysis Tools: CellRank 2’s advanced methods for identifying terminal states, calculating fate probabilities, and pinpointing driver genes offer researchers powerful tools for dissecting complex biological processes.
The impact of CellRank 2 extends beyond its immediate applications. By providing a standardized, flexible approach to trajectory inference, it has the potential to:
- Accelerate discoveries in developmental biology and regenerative medicine
- Enhance our understanding of disease progression, particularly in cancer research
- Facilitate the integration of emerging single-cell technologies into trajectory analysis workflows
- Foster collaboration and comparison across studies by providing a common analytical framework
As single-cell technologies continue to evolve, the modular design of CellRank 2 positions it to adapt and incorporate new data types and analytical methods. This adaptability ensures that CellRank 2 will remain a valuable tool in the single-cell biology toolkit for years to come.
In conclusion, CellRank 2 represents a significant leap forward in our ability to decipher the complex dynamics of cellular differentiation and fate decisions. Its combination of flexibility, performance, and analytical power makes it an invaluable resource for researchers seeking to unlock the secrets hidden within single-cell data. As we look to the future, CellRank 2 promises to play a crucial role in driving new discoveries and deepening our understanding of cellular biology at its most fundamental level.
This post was written with the help of Claude 3.5 Sonnet.

Leave a comment